Matthew R. Carbone
A Universal Deep Learning Framework for Materials X-ray Absorption Spectra
Sep 29, 2024Abstract:X-ray absorption spectroscopy (XAS) is a powerful characterization technique for probing the local chemical environment of absorbing atoms. However, analyzing XAS data presents with significant challenges, often requiring extensive, computationally intensive simulations, as well as significant domain expertise. These limitations hinder the development of fast, robust XAS analysis pipelines that are essential in high-throughput studies and for autonomous experimentation. We address these challenges with a suite of transfer learning approaches for XAS prediction, each uniquely contributing to improved accuracy and efficiency, as demonstrated on K-edge spectra database covering eight 3d transition metals (Ti-Cu). Our framework is built upon three distinct strategies. First, we use M3GNet to derive latent representations of the local chemical environment of absorption sites as input for XAS prediction, achieving up to order-of-magnitude improvements over conventional featurization techniques. Second, we employ a hierarchical transfer learning strategy, training a universal multi-task model across elements before fine-tuning for element-specific predictions. This cascaded approach after element-wise fine-turning yields models that outperform element-specific models by up to 31\%. Third, we implement cross-fidelity transfer learning, adapting a universal model to predict spectra generated by simulation of a different fidelity with a much higher computational cost. This approach improves prediction accuracy by up to 24\% over models trained on the target fidelity alone. Our approach is extendable to XAS prediction for a broader range of elements and offers a generalizable transfer learning framework to enhance other deep-learning models in materials science.
Spectroscopy-Guided Discovery of Three-Dimensional Structures of Disordered Materials with Diffusion Models
Dec 09, 2023



Abstract:The ability to rapidly develop materials with desired properties has a transformative impact on a broad range of emerging technologies. In this work, we introduce a new framework based on the diffusion model, a recent generative machine learning method to predict 3D structures of disordered materials from a target property. For demonstration, we apply the model to identify the atomic structures of amorphous carbons ($a$-C) as a representative material system from the target X-ray absorption near edge structure (XANES) spectra--a common experimental technique to probe atomic structures of materials. We show that conditional generation guided by XANES spectra reproduces key features of the target structures. Furthermore, we show that our model can steer the generative process to tailor atomic arrangements for a specific XANES spectrum. Finally, our generative model exhibits a remarkable scale-agnostic property, thereby enabling generation of realistic, large-scale structures through learning from a small-scale dataset (i.e., with small unit cells). Our work represents a significant stride in bridging the gap between materials characterization and atomic structure determination; in addition, it can be leveraged for materials discovery in exploring various material properties as targeted.
Transferable Graph Neural Fingerprint Models for Quick Response to Future Bio-Threats
Jul 17, 2023



Abstract:Fast screening of drug molecules based on the ligand binding affinity is an important step in the drug discovery pipeline. Graph neural fingerprint is a promising method for developing molecular docking surrogates with high throughput and great fidelity. In this study, we built a COVID-19 drug docking dataset of about 300,000 drug candidates on 23 coronavirus protein targets. With this dataset, we trained graph neural fingerprint docking models for high-throughput virtual COVID-19 drug screening. The graph neural fingerprint models yield high prediction accuracy on docking scores with the mean squared error lower than $0.21$ kcal/mol for most of the docking targets, showing significant improvement over conventional circular fingerprint methods. To make the neural fingerprints transferable for unknown targets, we also propose a transferable graph neural fingerprint method trained on multiple targets. With comparable accuracy to target-specific graph neural fingerprint models, the transferable model exhibits superb training and data efficiency. We highlight that the impact of this study extends beyond COVID-19 dataset, as our approach for fast virtual ligand screening can be easily adapted and integrated into a general machine learning-accelerated pipeline to battle future bio-threats.
Decoding Structure-Spectrum Relationships with Physically Organized Latent Spaces
Jan 11, 2023

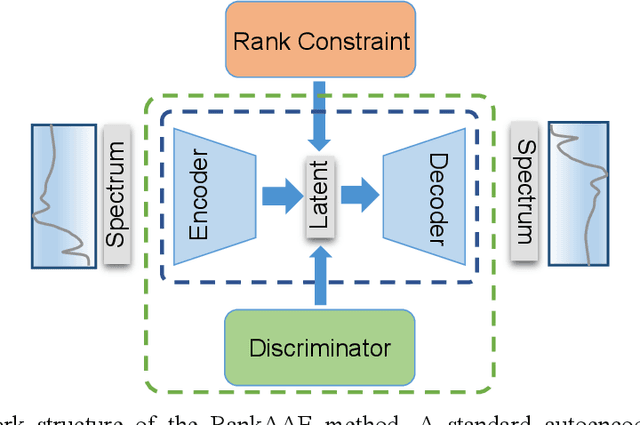

Abstract:A new semi-supervised machine learning method for the discovery of structure-spectrum relationships is developed and demonstrated using the specific example of interpreting X-ray absorption near-edge structure (XANES) spectra. This method constructs a one-to-one mapping between individual structure descriptors and spectral trends. Specifically, an adversarial autoencoder is augmented with a novel rank constraint (RankAAE). The RankAAE methodology produces a continuous and interpretable latent space, where each dimension can track an individual structure descriptor. As a part of this process, the model provides a robust and quantitative measure of the structure-spectrum relationship by decoupling intertwined spectral contributions from multiple structural characteristics. This makes it ideal for spectral interpretation and the discovery of new descriptors. The capability of this procedure is showcased by considering five local structure descriptors and a database of over fifty thousand simulated XANES spectra across eight first-row transition metal oxide families. The resulting structure-spectrum relationships not only reproduce known trends in the literature, but also reveal unintuitive ones that are visually indiscernible in large data sets. The results suggest that the RankAAE methodology has great potential to assist researchers to interpret complex scientific data, test physical hypotheses, and reveal new patterns that extend scientific insight.
Machine-learning Kondo physics using variational autoencoders
Jul 16, 2021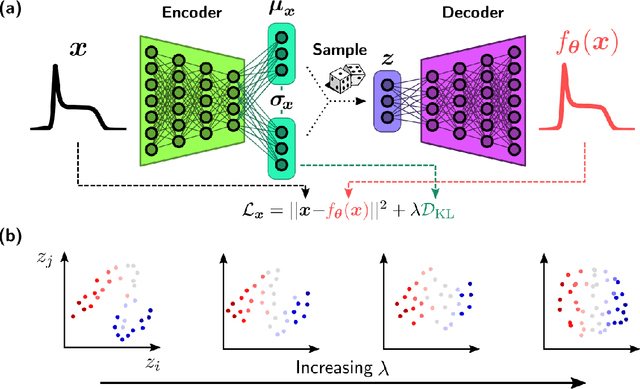
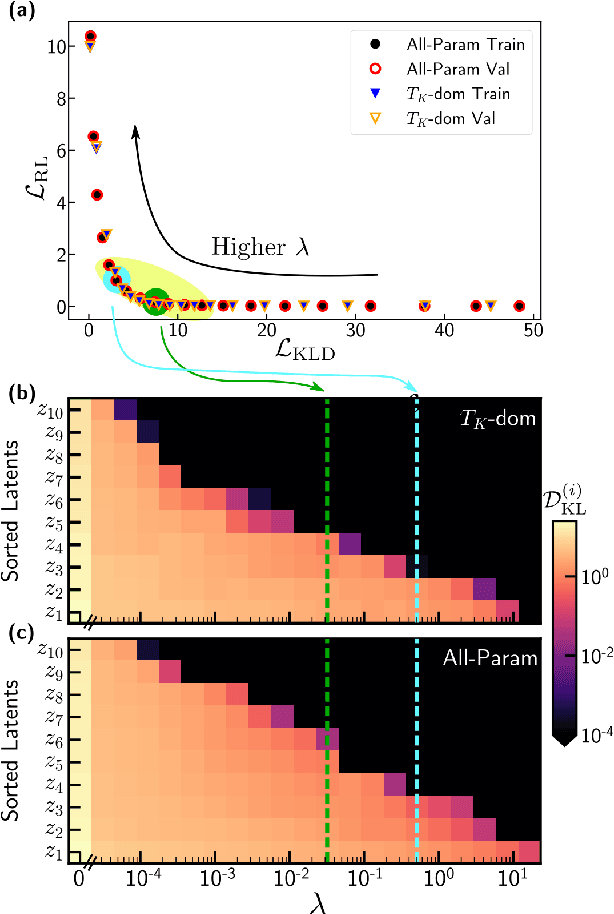
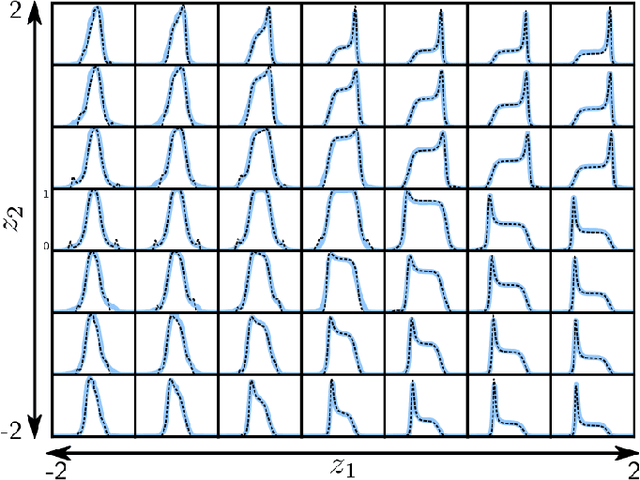
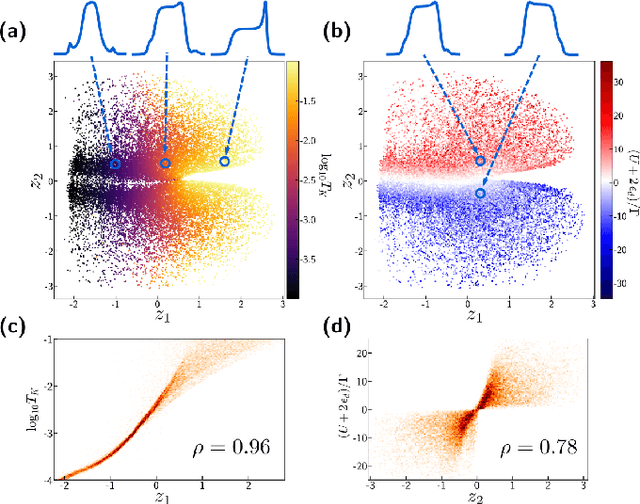
Abstract:We employ variational autoencoders to extract physical insight from a dataset of one-particle Anderson impurity model spectral functions. Autoencoders are trained to find a low-dimensional, latent space representation that faithfully characterizes each element of the training set, as measured by a reconstruction error. Variational autoencoders, a probabilistic generalization of standard autoencoders, further condition the learned latent space to promote highly interpretable features. In our study, we find that the learned latent space components strongly correlate with well known, but nontrivial, parameters that characterize emergent behaviors in the Anderson impurity model. In particular, one latent space component correlates with particle-hole asymmetry, while another is in near one-to-one correspondence with the Kondo temperature, a dynamically generated low-energy scale in the impurity model. With symbolic regression, we model this component as a function of bare physical input parameters and "rediscover" the non-perturbative formula for the Kondo temperature. The machine learning pipeline we develop opens opportunities to discover new domain knowledge in other physical systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge