Luc Soler
U-Net Transformer: Self and Cross Attention for Medical Image Segmentation
Mar 12, 2021



Abstract:Medical image segmentation remains particularly challenging for complex and low-contrast anatomical structures. In this paper, we introduce the U-Transformer network, which combines a U-shaped architecture for image segmentation with self- and cross-attention from Transformers. U-Transformer overcomes the inability of U-Nets to model long-range contextual interactions and spatial dependencies, which are arguably crucial for accurate segmentation in challenging contexts. To this end, attention mechanisms are incorporated at two main levels: a self-attention module leverages global interactions between encoder features, while cross-attention in the skip connections allows a fine spatial recovery in the U-Net decoder by filtering out non-semantic features. Experiments on two abdominal CT-image datasets show the large performance gain brought out by U-Transformer compared to U-Net and local Attention U-Nets. We also highlight the importance of using both self- and cross-attention, and the nice interpretability features brought out by U-Transformer.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019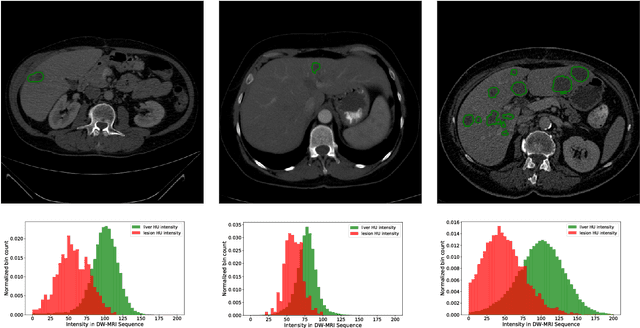
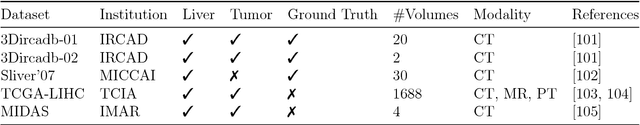
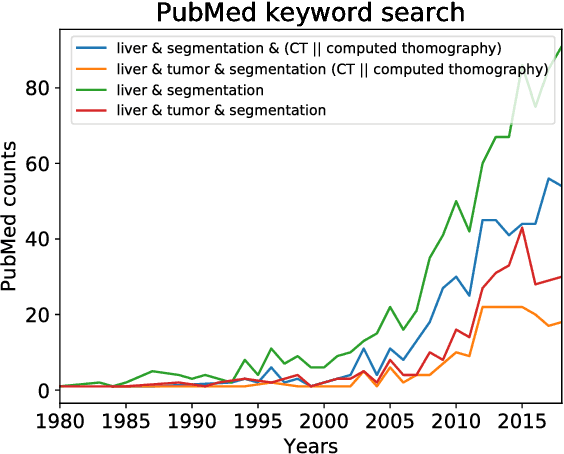
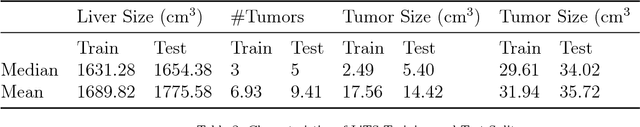
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
SLAM based Quasi Dense Reconstruction For Minimally Invasive Surgery Scenes
May 25, 2017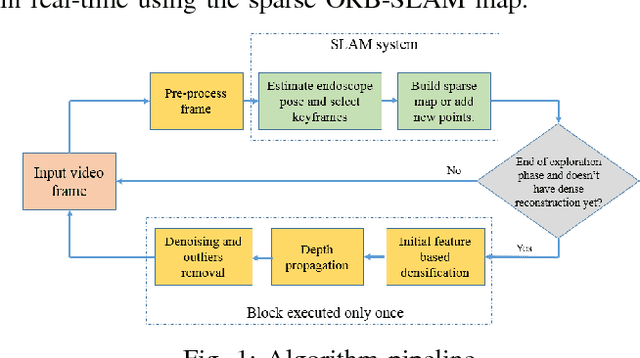
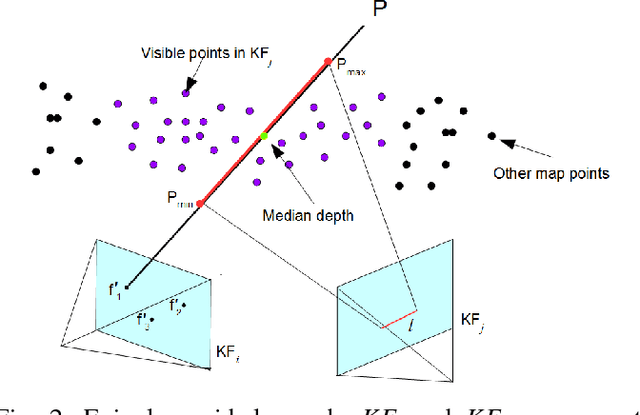
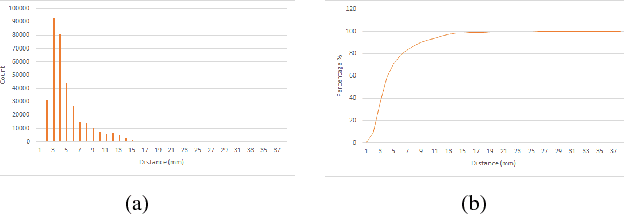
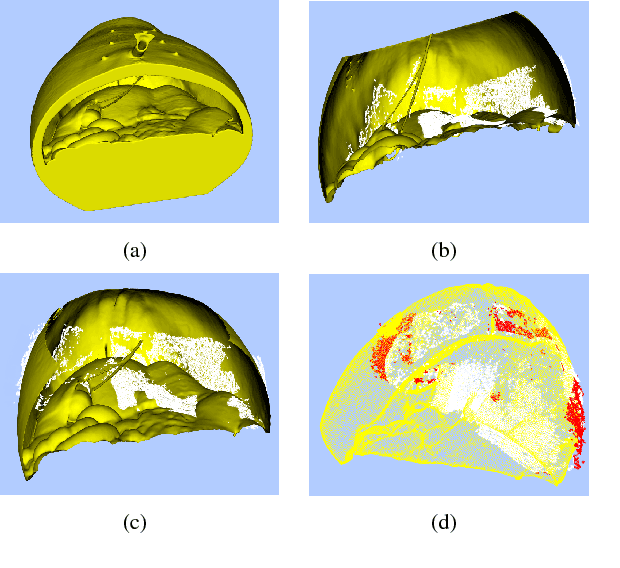
Abstract:Recovering surgical scene structure in laparoscope surgery is crucial step for surgical guidance and augmented reality applications. In this paper, a quasi dense reconstruction algorithm of surgical scene is proposed. This is based on a state-of-the-art SLAM system, and is exploiting the initial exploration phase that is typically performed by the surgeon at the beginning of the surgery. We show how to convert the sparse SLAM map to a quasi dense scene reconstruction, using pairs of keyframe images and correlation-based featureless patch matching. We have validated the approach with a live porcine experiment using Computed Tomography as ground truth, yielding a Root Mean Squared Error of 4.9mm.
Automatic View-Point Selection for Inter-Operative Endoscopic Surveillance
Oct 13, 2016



Abstract:Esophageal adenocarcinoma arises from Barrett's esophagus, which is the most serious complication of gastroesophageal reflux disease. Strategies for screening involve periodic surveillance and tissue biopsies. A major challenge in such regular examinations is to record and track the disease evolution and re-localization of biopsied sites to provide targeted treatments. In this paper, we extend our original inter-operative relocalization framework to provide a constrained image based search for obtaining the best view-point match to the live view. Within this context we investigate the effect of: the choice of feature descriptors and color-space; filtering of uninformative frames and endoscopic modality, for view-point localization. Our experiments indicate an improvement in the best view-point retrieval rate to [92%,87%] from [73%,76%] (in our previous approach) for NBI and WL.
ORBSLAM-based Endoscope Tracking and 3D Reconstruction
Aug 29, 2016
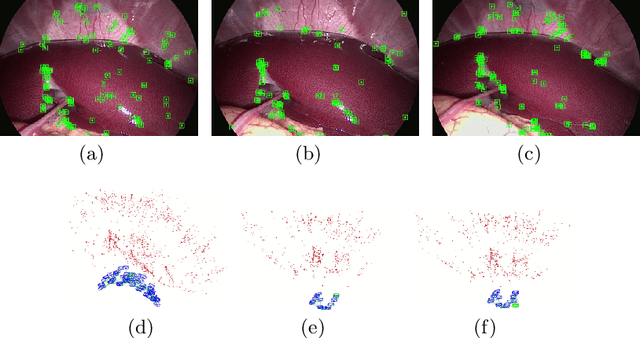
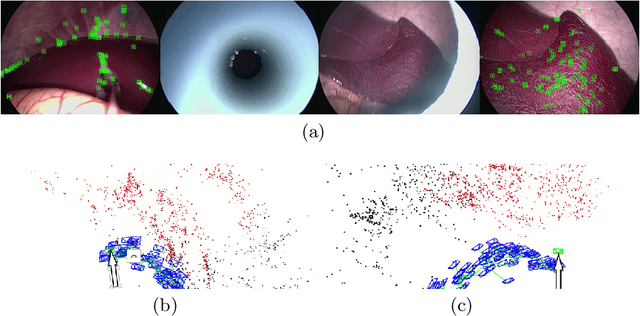
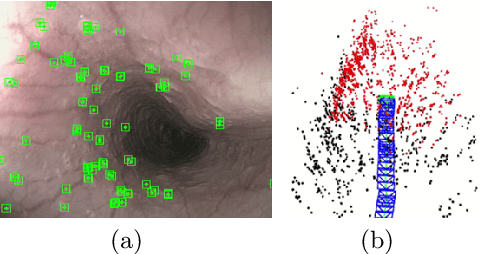
Abstract:We aim to track the endoscope location inside the surgical scene and provide 3D reconstruction, in real-time, from the sole input of the image sequence captured by the monocular endoscope. This information offers new possibilities for developing surgical navigation and augmented reality applications. The main benefit of this approach is the lack of extra tracking elements which can disturb the surgeon performance in the clinical routine. It is our first contribution to exploit ORBSLAM, one of the best performing monocular SLAM algorithms, to estimate both of the endoscope location, and 3D structure of the surgical scene. However, the reconstructed 3D map poorly describe textureless soft organ surfaces such as liver. It is our second contribution to extend ORBSLAM to be able to reconstruct a semi-dense map of soft organs. Experimental results on in-vivo pigs, shows a robust endoscope tracking even with organs deformations and partial instrument occlusions. It also shows the reconstruction density, and accuracy against ground truth surface obtained from CT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge