Alexandre Hostettler
TRUSTED: The Paired 3D Transabdominal Ultrasound and CT Human Data for Kidney Segmentation and Registration Research
Oct 19, 2023Abstract:Inter-modal image registration (IMIR) and image segmentation with abdominal Ultrasound (US) data has many important clinical applications, including image-guided surgery, automatic organ measurement and robotic navigation. However, research is severely limited by the lack of public datasets. We propose TRUSTED (the Tridimensional Renal Ultra Sound TomodEnsitometrie Dataset), comprising paired transabdominal 3DUS and CT kidney images from 48 human patients (96 kidneys), including segmentation, and anatomical landmark annotations by two experienced radiographers. Inter-rater segmentation agreement was over 94 (Dice score), and gold-standard segmentations were generated using the STAPLE algorithm. Seven anatomical landmarks were annotated, important for IMIR systems development and evaluation. To validate the dataset's utility, 5 competitive Deep Learning models for automatic kidney segmentation were benchmarked, yielding average DICE scores from 83.2% to 89.1% for CT, and 61.9% to 79.4% for US images. Three IMIR methods were benchmarked, and Coherent Point Drift performed best with an average Target Registration Error of 4.53mm. The TRUSTED dataset may be used freely researchers to develop and validate new segmentation and IMIR methods.
INDEXITY: a web-based collaborative tool for medical video annotation
Jun 26, 2023



Abstract:This technical report presents Indexity 1.4.0, a web-based tool designed for medical video annotation in surgical data science projects. We describe the main features available for the management of videos, annotations, ontology and users, as well as the global software architecture.
Full Contextual Attention for Multi-resolution Transformers in Semantic Segmentation
Dec 15, 2022



Abstract:Transformers have proved to be very effective for visual recognition tasks. In particular, vision transformers construct compressed global representations through self-attention and learnable class tokens. Multi-resolution transformers have shown recent successes in semantic segmentation but can only capture local interactions in high-resolution feature maps. This paper extends the notion of global tokens to build GLobal Attention Multi-resolution (GLAM) transformers. GLAM is a generic module that can be integrated into most existing transformer backbones. GLAM includes learnable global tokens, which unlike previous methods can model interactions between all image regions, and extracts powerful representations during training. Extensive experiments show that GLAM-Swin or GLAM-Swin-UNet exhibit substantially better performances than their vanilla counterparts on ADE20K and Cityscapes. Moreover, GLAM can be used to segment large 3D medical images, and GLAM-nnFormer achieves new state-of-the-art performance on the BCV dataset.
Memory transformers for full context and high-resolution 3D Medical Segmentation
Oct 11, 2022



Abstract:Transformer models achieve state-of-the-art results for image segmentation. However, achieving long-range attention, necessary to capture global context, with high-resolution 3D images is a fundamental challenge. This paper introduces the Full resolutIoN mEmory (FINE) transformer to overcome this issue. The core idea behind FINE is to learn memory tokens to indirectly model full range interactions while scaling well in both memory and computational costs. FINE introduces memory tokens at two levels: the first one allows full interaction between voxels within local image regions (patches), the second one allows full interactions between all regions of the 3D volume. Combined, they allow full attention over high resolution images, e.g. 512 x 512 x 256 voxels and above. Experiments on the BCV image segmentation dataset shows better performances than state-of-the-art CNN and transformer baselines, highlighting the superiority of our full attention mechanism compared to recent transformer baselines, e.g. CoTr, and nnFormer.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019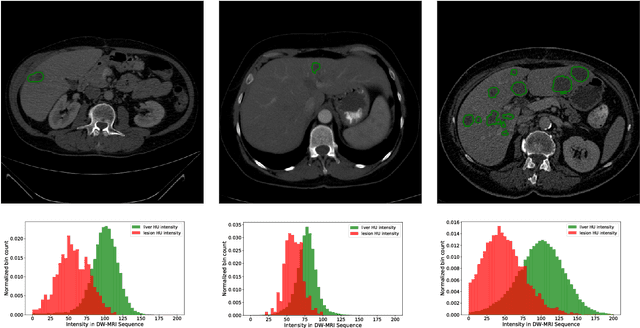
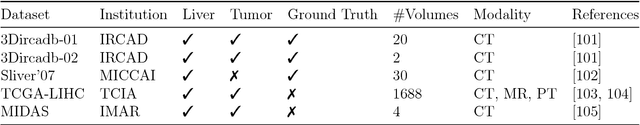
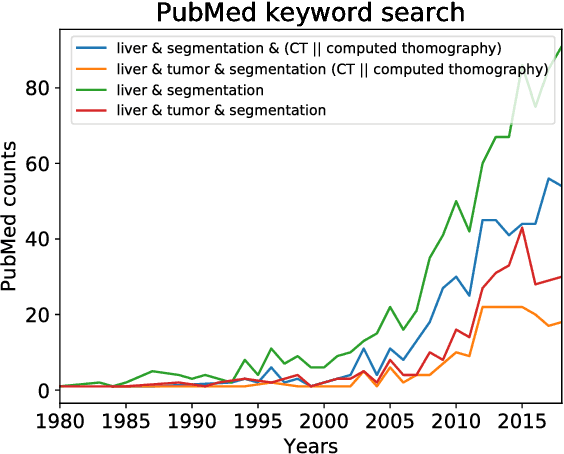
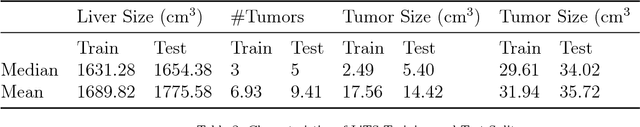
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
SLAM based Quasi Dense Reconstruction For Minimally Invasive Surgery Scenes
May 25, 2017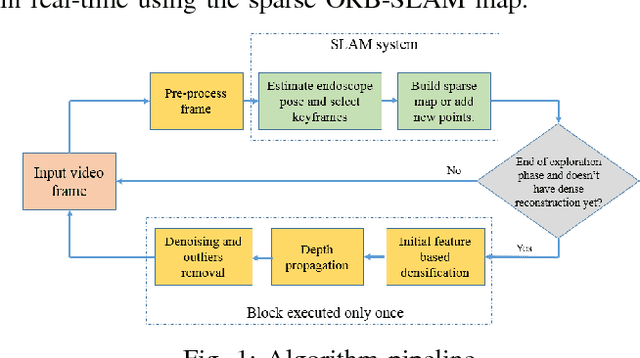
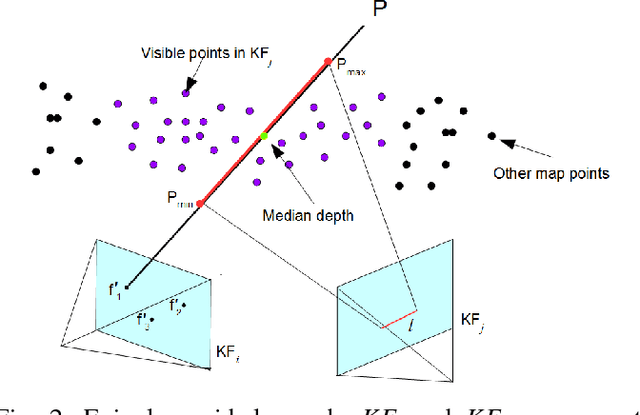
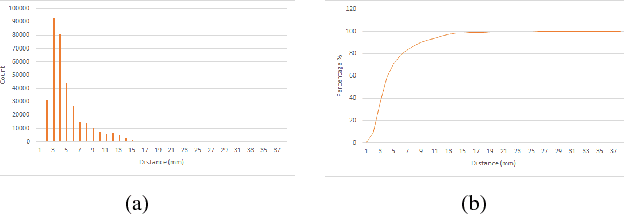
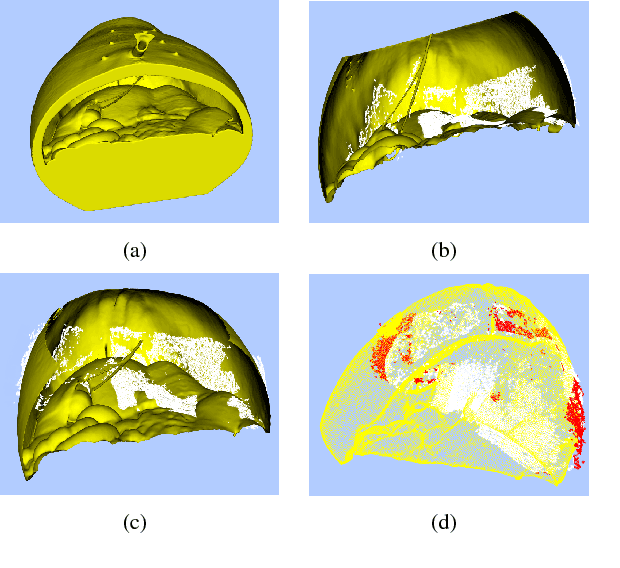
Abstract:Recovering surgical scene structure in laparoscope surgery is crucial step for surgical guidance and augmented reality applications. In this paper, a quasi dense reconstruction algorithm of surgical scene is proposed. This is based on a state-of-the-art SLAM system, and is exploiting the initial exploration phase that is typically performed by the surgeon at the beginning of the surgery. We show how to convert the sparse SLAM map to a quasi dense scene reconstruction, using pairs of keyframe images and correlation-based featureless patch matching. We have validated the approach with a live porcine experiment using Computed Tomography as ground truth, yielding a Root Mean Squared Error of 4.9mm.
ORBSLAM-based Endoscope Tracking and 3D Reconstruction
Aug 29, 2016
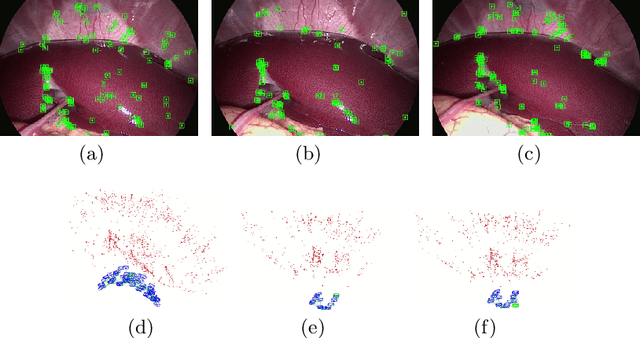
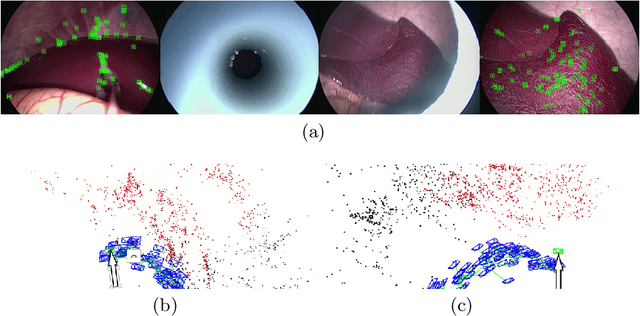
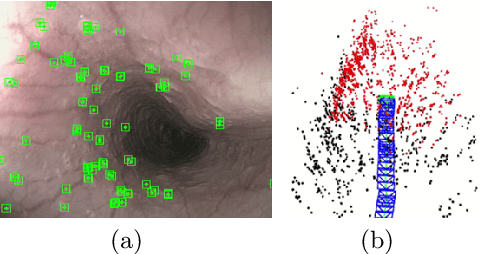
Abstract:We aim to track the endoscope location inside the surgical scene and provide 3D reconstruction, in real-time, from the sole input of the image sequence captured by the monocular endoscope. This information offers new possibilities for developing surgical navigation and augmented reality applications. The main benefit of this approach is the lack of extra tracking elements which can disturb the surgeon performance in the clinical routine. It is our first contribution to exploit ORBSLAM, one of the best performing monocular SLAM algorithms, to estimate both of the endoscope location, and 3D structure of the surgical scene. However, the reconstructed 3D map poorly describe textureless soft organ surfaces such as liver. It is our second contribution to extend ORBSLAM to be able to reconstruct a semi-dense map of soft organs. Experimental results on in-vivo pigs, shows a robust endoscope tracking even with organs deformations and partial instrument occlusions. It also shows the reconstruction density, and accuracy against ground truth surface obtained from CT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge