Lixing Shen
MHA2MLA-VLM: Enabling DeepSeek's Economical Multi-Head Latent Attention across Vision-Language Models
Jan 16, 2026Abstract:As vision-language models (VLMs) tackle increasingly complex and multimodal tasks, the rapid growth of Key-Value (KV) cache imposes significant memory and computational bottlenecks during inference. While Multi-Head Latent Attention (MLA) offers an effective means to compress the KV cache and accelerate inference, adapting existing VLMs to the MLA architecture without costly pretraining remains largely unexplored. In this work, we present MHA2MLA-VLM, a parameter-efficient and multimodal-aware framework for converting off-the-shelf VLMs to MLA. Our approach features two core techniques: (1) a modality-adaptive partial-RoPE strategy that supports both traditional and multimodal settings by selectively masking nonessential dimensions, and (2) a modality-decoupled low-rank approximation method that independently compresses the visual and textual KV spaces. Furthermore, we introduce parameter-efficient fine-tuning to minimize adaptation cost and demonstrate that minimizing output activation error, rather than parameter distance, substantially reduces performance loss. Extensive experiments on three representative VLMs show that MHA2MLA-VLM restores original model performance with minimal supervised data, significantly reduces KV cache footprint, and integrates seamlessly with KV quantization.
Towards Economical Inference: Enabling DeepSeek's Multi-Head Latent Attention in Any Transformer-based LLMs
Feb 20, 2025Abstract:Multi-head Latent Attention (MLA) is an innovative architecture proposed by DeepSeek, designed to ensure efficient and economical inference by significantly compressing the Key-Value (KV) cache into a latent vector. Compared to MLA, standard LLMs employing Multi-Head Attention (MHA) and its variants such as Grouped-Query Attention (GQA) exhibit significant cost disadvantages. Enabling well-trained LLMs (e.g., Llama) to rapidly adapt to MLA without pre-training from scratch is both meaningful and challenging. This paper proposes the first data-efficient fine-tuning method for transitioning from MHA to MLA (MHA2MLA), which includes two key components: for partial-RoPE, we remove RoPE from dimensions of queries and keys that contribute less to the attention scores, for low-rank approximation, we introduce joint SVD approximations based on the pre-trained parameters of keys and values. These carefully designed strategies enable MHA2MLA to recover performance using only a small fraction (0.3% to 0.6%) of the data, significantly reducing inference costs while seamlessly integrating with compression techniques such as KV cache quantization. For example, the KV cache size of Llama2-7B is reduced by 92.19%, with only a 0.5% drop in LongBench performance.
Secrets of RLHF in Large Language Models Part II: Reward Modeling
Jan 12, 2024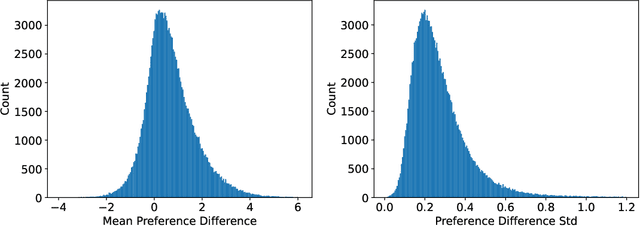

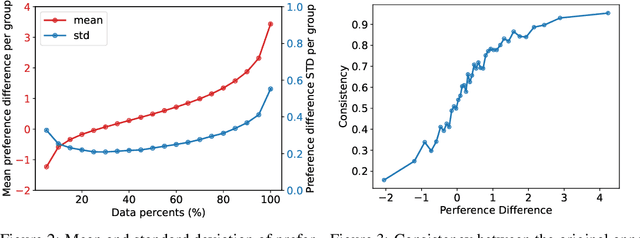
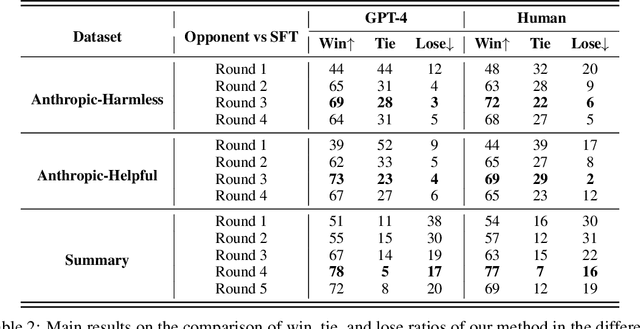
Abstract:Reinforcement Learning from Human Feedback (RLHF) has become a crucial technology for aligning language models with human values and intentions, enabling models to produce more helpful and harmless responses. Reward models are trained as proxies for human preferences to drive reinforcement learning optimization. While reward models are often considered central to achieving high performance, they face the following challenges in practical applications: (1) Incorrect and ambiguous preference pairs in the dataset may hinder the reward model from accurately capturing human intent. (2) Reward models trained on data from a specific distribution often struggle to generalize to examples outside that distribution and are not suitable for iterative RLHF training. In this report, we attempt to address these two issues. (1) From a data perspective, we propose a method to measure the strength of preferences within the data, based on a voting mechanism of multiple reward models. Experimental results confirm that data with varying preference strengths have different impacts on reward model performance. We introduce a series of novel methods to mitigate the influence of incorrect and ambiguous preferences in the dataset and fully leverage high-quality preference data. (2) From an algorithmic standpoint, we introduce contrastive learning to enhance the ability of reward models to distinguish between chosen and rejected responses, thereby improving model generalization. Furthermore, we employ meta-learning to enable the reward model to maintain the ability to differentiate subtle differences in out-of-distribution samples, and this approach can be utilized for iterative RLHF optimization.
Two Step Joint Model for Drug Drug Interaction Extraction
Aug 28, 2020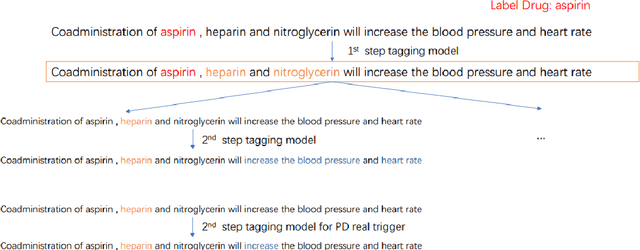
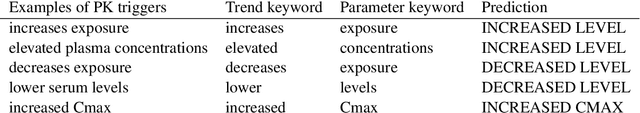
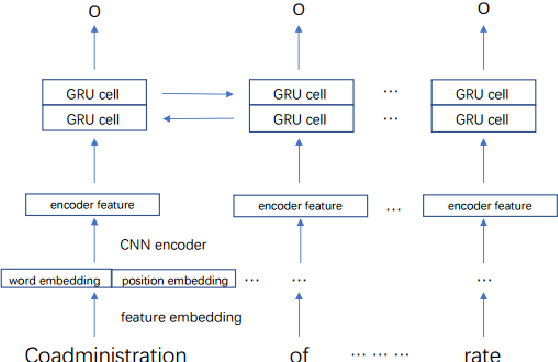

Abstract:When patients need to take medicine, particularly taking more than one kind of drug simultaneously, they should be alarmed that there possibly exists drug-drug interaction. Interaction between drugs may have a negative impact on patients or even cause death. Generally, drugs that conflict with a specific drug (or label drug) are usually described in its drug label or package insert. Since more and more new drug products come into the market, it is difficult to collect such information by manual. We take part in the Drug-Drug Interaction (DDI) Extraction from Drug Labels challenge of Text Analysis Conference (TAC) 2018, choosing task1 and task2 to automatically extract DDI related mentions and DDI relations respectively. Instead of regarding task1 as named entity recognition (NER) task and regarding task2 as relation extraction (RE) task then solving it in a pipeline, we propose a two step joint model to detect DDI and it's related mentions jointly. A sequence tagging system (CNN-GRU encoder-decoder) finds precipitants first and search its fine-grained Trigger and determine the DDI for each precipitant in the second step. Moreover, a rule based model is built to determine the sub-type for pharmacokinetic interation. Our system achieved best result in both task1 and task2. F-measure reaches 0.46 in task1 and 0.40 in task2.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge