Kin Long Kelvin Lee
Energy & Force Regression on DFT Trajectories is Not Enough for Universal Machine Learning Interatomic Potentials
Feb 05, 2025



Abstract:Universal Machine Learning Interactomic Potentials (MLIPs) enable accelerated simulations for materials discovery. However, current research efforts fail to impactfully utilize MLIPs due to: 1. Overreliance on Density Functional Theory (DFT) for MLIP training data creation; 2. MLIPs' inability to reliably and accurately perform large-scale molecular dynamics (MD) simulations for diverse materials; 3. Limited understanding of MLIPs' underlying capabilities. To address these shortcomings, we aargue that MLIP research efforts should prioritize: 1. Employing more accurate simulation methods for large-scale MLIP training data creation (e.g. Coupled Cluster Theory) that cover a wide range of materials design spaces; 2. Creating MLIP metrology tools that leverage large-scale benchmarking, visualization, and interpretability analyses to provide a deeper understanding of MLIPs' inner workings; 3. Developing computationally efficient MLIPs to execute MD simulations that accurately model a broad set of materials properties. Together, these interdisciplinary research directions can help further the real-world application of MLIPs to accurately model complex materials at device scale.
SymmCD: Symmetry-Preserving Crystal Generation with Diffusion Models
Feb 05, 2025Abstract:Generating novel crystalline materials has potential to lead to advancements in fields such as electronics, energy storage, and catalysis. The defining characteristic of crystals is their symmetry, which plays a central role in determining their physical properties. However, existing crystal generation methods either fail to generate materials that display the symmetries of real-world crystals, or simply replicate the symmetry information from examples in a database. To address this limitation, we propose SymmCD, a novel diffusion-based generative model that explicitly incorporates crystallographic symmetry into the generative process. We decompose crystals into two components and learn their joint distribution through diffusion: 1) the asymmetric unit, the smallest subset of the crystal which can generate the whole crystal through symmetry transformations, and; 2) the symmetry transformations needed to be applied to each atom in the asymmetric unit. We also use a novel and interpretable representation for these transformations, enabling generalization across different crystallographic symmetry groups. We showcase the competitive performance of SymmCD on a subset of the Materials Project, obtaining diverse and valid crystals with realistic symmetries and predicted properties.
Stiefel Flow Matching for Moment-Constrained Structure Elucidation
Dec 17, 2024



Abstract:Molecular structure elucidation is a fundamental step in understanding chemical phenomena, with applications in identifying molecules in natural products, lab syntheses, forensic samples, and the interstellar medium. We consider the task of predicting a molecule's all-atom 3D structure given only its molecular formula and moments of inertia, motivated by the ability of rotational spectroscopy to measure these moments. While existing generative models can conditionally sample 3D structures with approximately correct moments, this soft conditioning fails to leverage the many digits of precision afforded by experimental rotational spectroscopy. To address this, we first show that the space of $n$-atom point clouds with a fixed set of moments of inertia is embedded in the Stiefel manifold $\mathrm{St}(n, 4)$. We then propose Stiefel Flow Matching as a generative model for elucidating 3D structure under exact moment constraints. Additionally, we learn simpler and shorter flows by finding approximate solutions for equivariant optimal transport on the Stiefel manifold. Empirically, enforcing exact moment constraints allows Stiefel Flow Matching to achieve higher success rates and faster sampling than Euclidean diffusion models, even on high-dimensional manifolds corresponding to large molecules in the GEOM dataset.
Accelerating Quantum Emitter Characterization with Latent Neural Ordinary Differential Equations
Nov 17, 2024

Abstract:Deep neural network models can be used to learn complex dynamics from data and reconstruct sparse or noisy signals, thereby accelerating and augmenting experimental measurements. Evaluating the quantum optical properties of solid-state single-photon emitters is a time-consuming task that typically requires interferometric photon correlation experiments, such as Photon correlation Fourier spectroscopy (PCFS) which measures time-resolved single emitter lineshapes. Here, we demonstrate a latent neural ordinary differential equation model that can forecast a complete and noise-free PCFS experiment from a small subset of noisy correlation functions. By encoding measured photon correlations into an initial value problem, the NODE can be propagated to an arbitrary number of interferometer delay times. We demonstrate this with 10 noisy photon correlation functions that are used to extrapolate an entire de-noised interferograms of up to 200 stage positions, enabling up to a 20-fold speedup in experimental acquisition time from $\sim$3 hours to 10 minutes. Our work presents a new approach to greatly accelerate the experimental characterization of novel quantum emitter materials using deep learning.
Deconstructing equivariant representations in molecular systems
Oct 10, 2024Abstract:Recent equivariant models have shown significant progress in not just chemical property prediction, but as surrogates for dynamical simulations of molecules and materials. Many of the top performing models in this category are built within the framework of tensor products, which preserves equivariance by restricting interactions and transformations to those that are allowed by symmetry selection rules. Despite being a core part of the modeling process, there has not yet been much attention into understanding what information persists in these equivariant representations, and their general behavior outside of benchmark metrics. In this work, we report on a set of experiments using a simple equivariant graph convolution model on the QM9 dataset, focusing on correlating quantitative performance with the resulting molecular graph embeddings. Our key finding is that, for a scalar prediction task, many of the irreducible representations are simply ignored during training -- specifically those pertaining to vector ($l=1$) and tensor quantities ($l=2$) -- an issue that does not necessarily make itself evident in the test metric. We empirically show that removing some unused orders of spherical harmonics improves model performance, correlating with improved latent space structure. We provide a number of recommendations for future experiments to try and improve efficiency and utilization of equivariant features based on these observations.
$ρ$-Diffusion: A diffusion-based density estimation framework for computational physics
Dec 13, 2023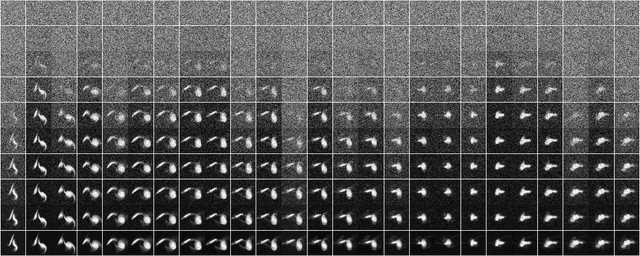

Abstract:In physics, density $\rho(\cdot)$ is a fundamentally important scalar function to model, since it describes a scalar field or a probability density function that governs a physical process. Modeling $\rho(\cdot)$ typically scales poorly with parameter space, however, and quickly becomes prohibitively difficult and computationally expensive. One promising avenue to bypass this is to leverage the capabilities of denoising diffusion models often used in high-fidelity image generation to parameterize $\rho(\cdot)$ from existing scientific data, from which new samples can be trivially sampled from. In this paper, we propose $\rho$-Diffusion, an implementation of denoising diffusion probabilistic models for multidimensional density estimation in physics, which is currently in active development and, from our results, performs well on physically motivated 2D and 3D density functions. Moreover, we propose a novel hashing technique that allows $\rho$-Diffusion to be conditioned by arbitrary amounts of physical parameters of interest.
MatSciML: A Broad, Multi-Task Benchmark for Solid-State Materials Modeling
Sep 12, 2023



Abstract:We propose MatSci ML, a novel benchmark for modeling MATerials SCIence using Machine Learning (MatSci ML) methods focused on solid-state materials with periodic crystal structures. Applying machine learning methods to solid-state materials is a nascent field with substantial fragmentation largely driven by the great variety of datasets used to develop machine learning models. This fragmentation makes comparing the performance and generalizability of different methods difficult, thereby hindering overall research progress in the field. Building on top of open-source datasets, including large-scale datasets like the OpenCatalyst, OQMD, NOMAD, the Carolina Materials Database, and Materials Project, the MatSci ML benchmark provides a diverse set of materials systems and properties data for model training and evaluation, including simulated energies, atomic forces, material bandgaps, as well as classification data for crystal symmetries via space groups. The diversity of properties in MatSci ML makes the implementation and evaluation of multi-task learning algorithms for solid-state materials possible, while the diversity of datasets facilitates the development of new, more generalized algorithms and methods across multiple datasets. In the multi-dataset learning setting, MatSci ML enables researchers to combine observations from multiple datasets to perform joint prediction of common properties, such as energy and forces. Using MatSci ML, we evaluate the performance of different graph neural networks and equivariant point cloud networks on several benchmark tasks spanning single task, multitask, and multi-data learning scenarios. Our open-source code is available at https://github.com/IntelLabs/matsciml.
The Open MatSci ML Toolkit: A Flexible Framework for Machine Learning in Materials Science
Oct 31, 2022



Abstract:We present the Open MatSci ML Toolkit: a flexible, self-contained, and scalable Python-based framework to apply deep learning models and methods on scientific data with a specific focus on materials science and the OpenCatalyst Dataset. Our toolkit provides: 1. A scalable machine learning workflow for materials science leveraging PyTorch Lightning, which enables seamless scaling across different computation capabilities (laptop, server, cluster) and hardware platforms (CPU, GPU, XPU). 2. Deep Graph Library (DGL) support for rapid graph neural network prototyping and development. By publishing and sharing this toolkit with the research community via open-source release, we hope to: 1. Lower the entry barrier for new machine learning researchers and practitioners that want to get started with the OpenCatalyst dataset, which presently comprises the largest computational materials science dataset. 2. Enable the scientific community to apply advanced machine learning tools to high-impact scientific challenges, such as modeling of materials behavior for clean energy applications. We demonstrate the capabilities of our framework by enabling three new equivariant neural network models for multiple OpenCatalyst tasks and arrive at promising results for compute scaling and model performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge