Jinpeng Jiang
Look Back for More: Harnessing Historical Sequential Updates for Personalized Federated Adapter Tuning
Jan 03, 2025



Abstract:Personalized federated learning (PFL) studies effective model personalization to address the data heterogeneity issue among clients in traditional federated learning (FL). Existing PFL approaches mainly generate personalized models by relying solely on the clients' latest updated models while ignoring their previous updates, which may result in suboptimal personalized model learning. To bridge this gap, we propose a novel framework termed pFedSeq, designed for personalizing adapters to fine-tune a foundation model in FL. In pFedSeq, the server maintains and trains a sequential learner, which processes a sequence of past adapter updates from clients and generates calibrations for personalized adapters. To effectively capture the cross-client and cross-step relations hidden in previous updates and generate high-performing personalized adapters, pFedSeq adopts the powerful selective state space model (SSM) as the architecture of sequential learner. Through extensive experiments on four public benchmark datasets, we demonstrate the superiority of pFedSeq over state-of-the-art PFL methods.
FedSSO: A Federated Server-Side Second-Order Optimization Algorithm
Jun 20, 2022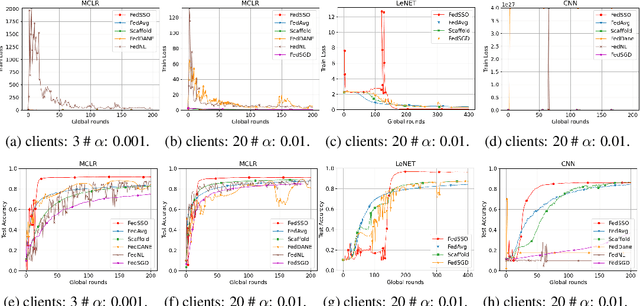
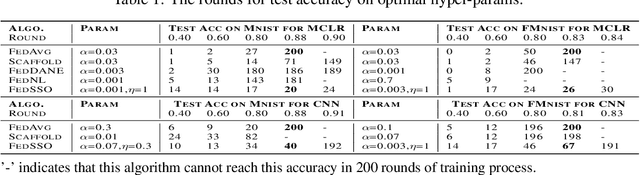
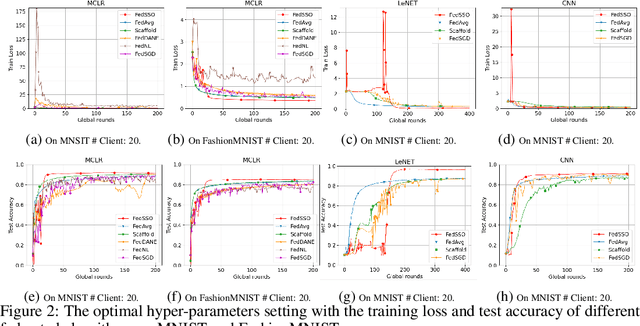
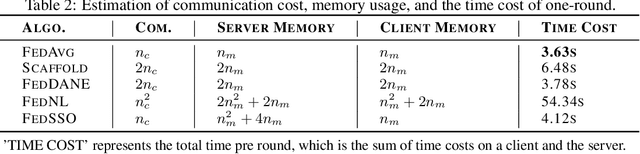
Abstract:In this work, we propose FedSSO, a server-side second-order optimization method for federated learning (FL). In contrast to previous works in this direction, we employ a server-side approximation for the Quasi-Newton method without requiring any training data from the clients. In this way, we not only shift the computation burden from clients to server, but also eliminate the additional communication for second-order updates between clients and server entirely. We provide theoretical guarantee for convergence of our novel method, and empirically demonstrate our fast convergence and communication savings in both convex and non-convex settings.
PrTransH: Embedding Probabilistic Medical Knowledge from Real World EMR Data
Sep 02, 2019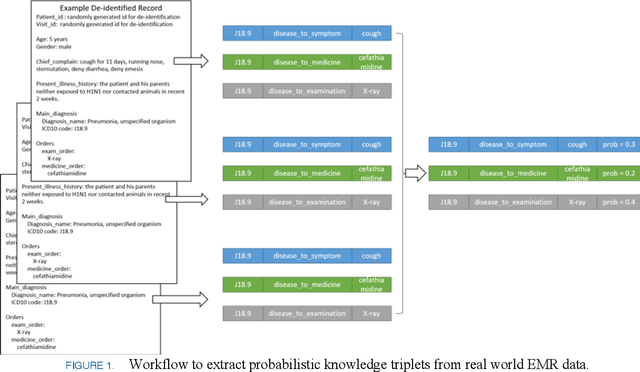
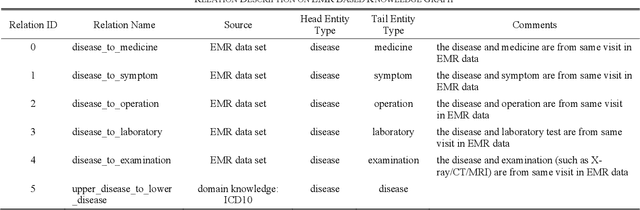
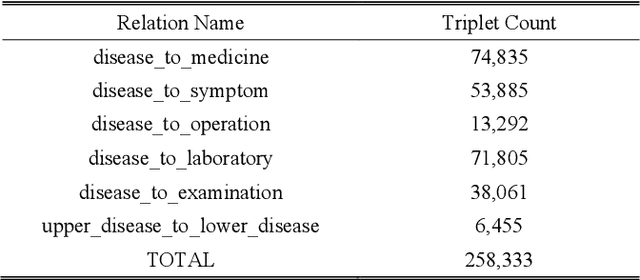
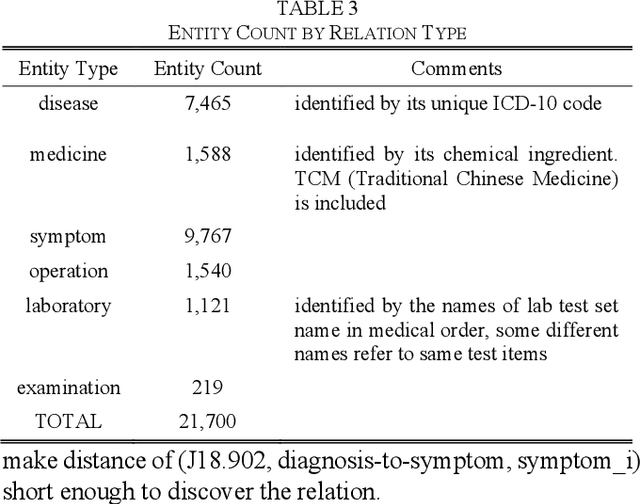
Abstract:This paper proposes an algorithm named as PrTransH to learn embedding vectors from real world EMR data based medical knowledge. The unique challenge in embedding medical knowledge graph from real world EMR data is that the uncertainty of knowledge triplets blurs the border between "correct triplet" and "wrong triplet", changing the fundamental assumption of many existing algorithms. To address the challenge, some enhancements are made to existing TransH algorithm, including: 1) involve probability of medical knowledge triplet into training objective; 2) replace the margin-based ranking loss with unified loss calculation considering both valid and corrupted triplets; 3) augment training data set with medical background knowledge. Verifications on real world EMR data based medical knowledge graph prove that PrTransH outperforms TransH in link prediction task. To the best of our survey, this paper is the first one to learn and verify knowledge embedding on probabilistic knowledge graphs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge