Jinjiang Guo
Predicting Molecule-Target Interaction by Learning Biomedical Network and Molecule Representations
Feb 06, 2023Abstract:The study of molecule-target interaction is quite important for drug discovery in terms of target identification, hit identification, pathway study, drug-drug interaction, etc. Most existing methodologies utilize either biomedical network information or molecule structural features to predict potential interaction link. However, the biomedical network information based methods usually suffer from cold start problem, while structure based methods often give limited performance due to the structure/interaction assumption and data quality. To address these issues, we propose a pseudo-siamese Graph Neural Network method, namely MTINet+, which learns both biomedical network topological and molecule structural/chemical information as representations to predict potential interaction of given molecule and target pair. In MTINet+, 1-hop subgraphs of given molecule and target pair are extracted from known interaction of biomedical network as topological information, meanwhile the molecule structural and chemical attributes are processed as molecule information. MTINet+ learns these two types of information as embedding features for predicting the pair link. In the experiments of different molecule-target interaction tasks, MTINet+ significantly outperforms over the state-of-the-art baselines. In addition, in our designed network sparsity experiments , MTINet+ shows strong robustness against different sparse biomedical networks.
Ligandformer: A Graph Neural Network for Predicting Compound Property with Robust Interpretation
Feb 24, 2022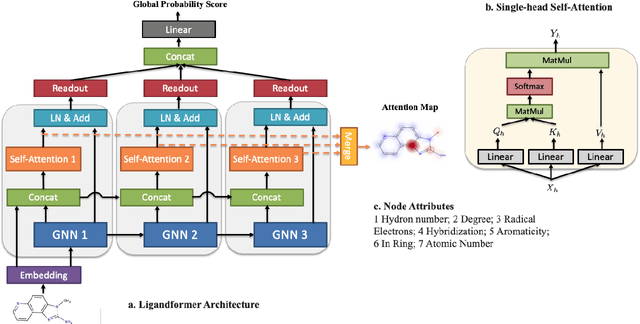

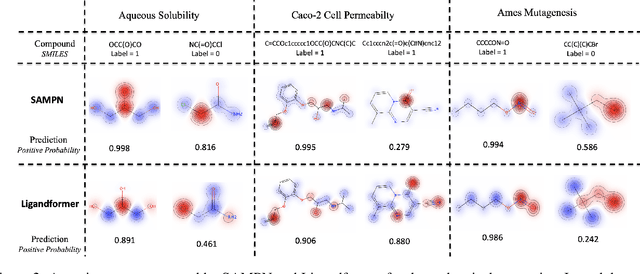
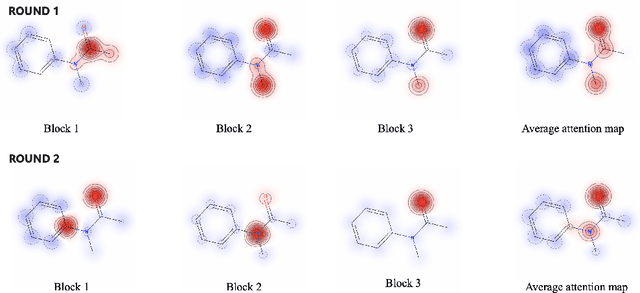
Abstract:Robust and efficient interpretation of QSAR methods is quite useful to validate AI prediction rationales with subjective opinion (chemist or biologist expertise), understand sophisticated chemical or biological process mechanisms, and provide heuristic ideas for structure optimization in pharmaceutical industry. For this purpose, we construct a multi-layer self-attention based Graph Neural Network framework, namely Ligandformer, for predicting compound property with interpretation. Ligandformer integrates attention maps on compound structure from different network blocks. The integrated attention map reflects the machine's local interest on compound structure, and indicates the relationship between predicted compound property and its structure. This work mainly contributes to three aspects: 1. Ligandformer directly opens the black-box of deep learning methods, providing local prediction rationales on chemical structures. 2. Ligandformer gives robust prediction in different experimental rounds, overcoming the ubiquitous prediction instability of deep learning methods. 3. Ligandformer can be generalized to predict different chemical or biological properties with high performance. Furthermore, Ligandformer can simultaneously output specific property score and visible attention map on structure, which can support researchers to investigate chemical or biological property and optimize structure efficiently. Our framework outperforms over counterparts in terms of accuracy, robustness and generalization, and can be applied in complex system study.
Heterogeneous Graph based Deep Learning for Biomedical Network Link Prediction
Feb 24, 2021



Abstract:Multi-scale biomedical knowledge networks are expanding with emerging experimental technologies that generates multi-scale biomedical big data. Link prediction is increasingly used especially in bipartite biomedical networks to identify hidden biological interactions and relationshipts between key entities such as compounds, targets, gene and diseases. We propose a Graph Neural Networks (GNN) method, namely Graph Pair based Link Prediction model (GPLP), for predicting biomedical network links simply based on their topological interaction information. In GPLP, 1-hop subgraphs extracted from known network interaction matrix is learnt to predict missing links. To evaluate our method, three heterogeneous biomedical networks were used, i.e. Drug-Target Interaction network (DTI), Compound-Protein Interaction network (CPI) from NIH Tox21, and Compound-Virus Inhibition network (CVI). Our proposed GPLP method significantly outperforms over the state-of-the-art baselines. In addition, different network incompleteness is analysed with our devised protocol, and we also design an effective approach to improve the model robustness towards incomplete networks. Our method demonstrates the potential applications in other biomedical networks.
Sequence-based deep learning antibody design for in silico antibody affinity maturation
Feb 21, 2021



Abstract:Antibody therapeutics has been extensively studied in drug discovery and development within the past decades. One increasingly popular focus in the antibody discovery pipeline is the optimization step for therapeutic leads. Both traditional methods and in silico approaches aim to generate candidates with high binding affinity against specific target antigens. Traditional in vitro approaches use hybridoma or phage display for candidate selection, and surface plasmon resonance (SPR) for evaluation, while in silico computational approaches aim to reduce the high cost and improve efficiency by incorporating mathematical algorithms and computational processing power in the design process. In the present study, we investigated different graph-based designs for depicting antibody-antigen interactions in terms of antibody affinity prediction using deep learning techniques. While other in silico computations require experimentally determined crystal structures, our study took interest in the capability of sequence-based models for in silico antibody maturation. Our preliminary studies achieved satisfying prediction accuracy on binding affinities comparing to conventional approaches and other deep learning approaches. To further study the antibody-antigen binding specificity, and to simulate the optimization process in real-world scenario, we introduced pairwise prediction strategy. We performed analysis based on both baseline and pairwise prediction results. The resulting prediction and efficiency prove the feasibility and computational efficiency of sequence-based method to be adapted as a scalable industry practice.
Real-time tracking of COVID-19 and coronavirus research updates through text mining
Feb 09, 2021



Abstract:The novel coronavirus (SARS-CoV-2) which causes COVID-19 is an ongoing pandemic. There are ongoing studies with up to hundreds of publications uploaded to databases daily. We are exploring the use-case of artificial intelligence and natural language processing in order to efficiently sort through these publications. We demonstrate that clinical trial information, preclinical studies, and a general topic model can be used as text mining data intelligence tools for scientists all over the world to use as a resource for their own research. To evaluate our method, several metrics are used to measure the information extraction and clustering results. In addition, we demonstrate that our workflow not only have a use-case for COVID-19, but for other disease areas as well. Overall, our system aims to allow scientists to more efficiently research coronavirus. Our automatically updating modules are available on our information portal at https://ghddi-ailab.github.io/Targeting2019-nCoV/ for public viewing.
Enhance Information Propagation for Graph Neural Network by Heterogeneous Aggregations
Feb 08, 2021



Abstract:Graph neural networks are emerging as continuation of deep learning success w.r.t. graph data. Tens of different graph neural network variants have been proposed, most following a neighborhood aggregation scheme, where the node features are updated via aggregating features of its neighboring nodes from layer to layer. Though related research surges, the power of GNNs are still not on-par-with their counterpart CNNs in computer vision and RNNs in natural language processing. We rethink this problem from the perspective of information propagation, and propose to enhance information propagation among GNN layers by combining heterogeneous aggregations. We argue that as richer information are propagated from shallow to deep layers, the discriminative capability of features formulated by GNN can benefit from it. As our first attempt in this direction, a new generic GNN layer formulation and upon this a new GNN variant referred as HAG-Net is proposed. We empirically validate the effectiveness of HAG-Net on a number of graph classification benchmarks, and elaborate all the design options and criterions along with.
Subjective and Objective Visual Quality Assessment of Textured 3D Meshes
Feb 08, 2021
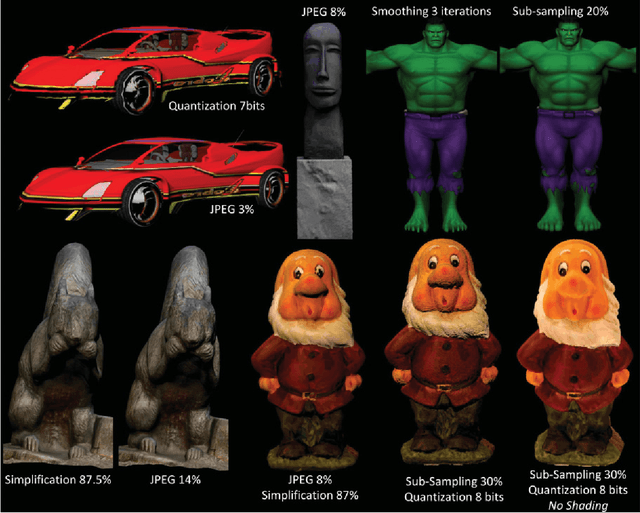

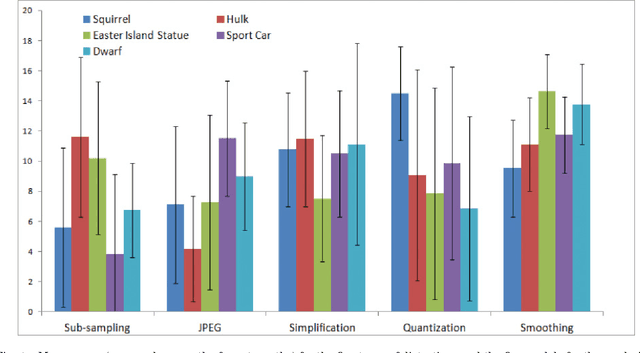
Abstract:Objective visual quality assessment of 3D models is a fundamental issue in computer graphics. Quality assessment metrics may allow a wide range of processes to be guided and evaluated, such as level of detail creation, compression, filtering, and so on. Most computer graphics assets are composed of geometric surfaces on which several texture images can be mapped to 11 make the rendering more realistic. While some quality assessment metrics exist for geometric surfaces, almost no research has been conducted on the evaluation of texture-mapped 3D models. In this context, we present a new subjective study to evaluate the perceptual quality of textured meshes, based on a paired comparison protocol. We introduce both texture and geometry distortions on a set of 5 reference models to produce a database of 136 distorted models, evaluated using two rendering protocols. Based on analysis of the results, we propose two new metrics for visual quality assessment of textured mesh, as optimized linear combinations of accurate geometry and texture quality measurements. These proposed perceptual metrics outperform their counterparts in terms of correlation with human opinion. The database, along with the associated subjective scores, will be made publicly available online.
Locally Adaptive Learning Loss for Semantic Image Segmentation
Apr 16, 2018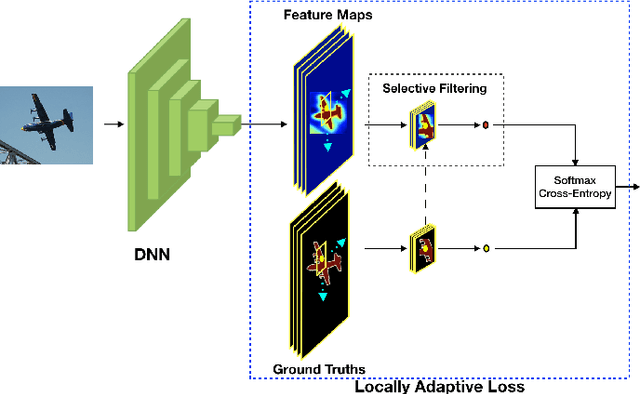

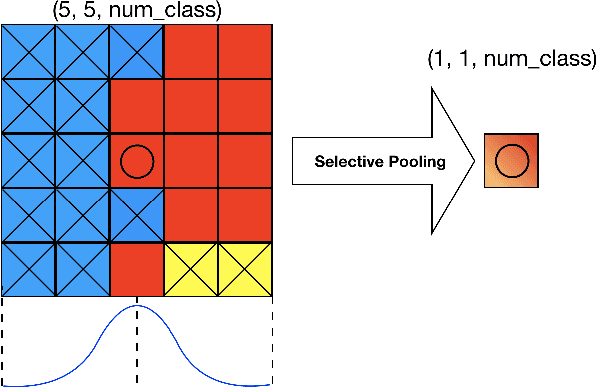
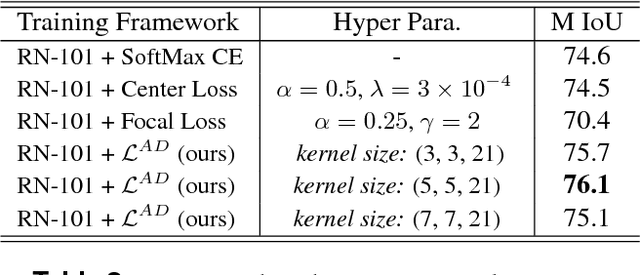
Abstract:We propose a novel locally adaptive learning estimator for enhancing the inter- and intra- discriminative capabilities of Deep Neural Networks, which can be used as improved loss layer for semantic image segmentation tasks. Most loss layers compute pixel-wise cost between feature maps and ground truths, ignoring spatial layouts and interactions between neighboring pixels with same object category, and thus networks cannot be effectively sensitive to intra-class connections. Stride by stride, our method firstly conducts adaptive pooling filter operating over predicted feature maps, aiming to merge predicted distributions over a small group of neighboring pixels with same category, and then it computes cost between the merged distribution vector and their category label. Such design can make groups of neighboring predictions from same category involved into estimations on predicting correctness with respect to their category, and hence train networks to be more sensitive to regional connections between adjacent pixels based on their categories. In the experiments on Pascal VOC 2012 segmentation datasets, the consistently improved results show that our proposed approach achieves better segmentation masks against previous counterparts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge