Jiarun Liu
TIGaussian: Disentangle Gaussians for Spatial-Awared Text-Image-3D Alignment
Jan 27, 2026Abstract:While visual-language models have profoundly linked features between texts and images, the incorporation of 3D modality data, such as point clouds and 3D Gaussians, further enables pretraining for 3D-related tasks, e.g., cross-modal retrieval, zero-shot classification, and scene recognition. As challenges remain in extracting 3D modal features and bridging the gap between different modalities, we propose TIGaussian, a framework that harnesses 3D Gaussian Splatting (3DGS) characteristics to strengthen cross-modality alignment through multi-branch 3DGS tokenizer and modality-specific 3D feature alignment strategies. Specifically, our multi-branch 3DGS tokenizer decouples the intrinsic properties of 3DGS structures into compact latent representations, enabling more generalizable feature extraction. To further bridge the modality gap, we develop a bidirectional cross-modal alignment strategies: a multi-view feature fusion mechanism that leverages diffusion priors to resolve perspective ambiguity in image-3D alignment, while a text-3D projection module adaptively maps 3D features to text embedding space for better text-3D alignment. Extensive experiments on various datasets demonstrate the state-of-the-art performance of TIGaussian in multiple tasks.
Unifying Dynamic Tool Creation and Cross-Task Experience Sharing through Cognitive Memory Architecture
Dec 12, 2025Abstract:Large Language Model agents face fundamental challenges in adapting to novel tasks due to limitations in tool availability and experience reuse. Existing approaches either rely on predefined tools with limited coverage or build tools from scratch without leveraging past experiences, leading to inefficient exploration and suboptimal performance. We introduce SMITH (Shared Memory Integrated Tool Hub), a unified cognitive architecture that seamlessly integrates dynamic tool creation with cross-task experience sharing through hierarchical memory organization. SMITH organizes agent memory into procedural, semantic, and episodic components, enabling systematic capability expansion while preserving successful execution patterns. Our approach formalizes tool creation as iterative code generation within controlled sandbox environments and experience sharing through episodic memory retrieval with semantic similarity matching. We further propose a curriculum learning strategy based on agent-ensemble difficulty re-estimation. Extensive experiments on the GAIA benchmark demonstrate SMITH's effectiveness, achieving 81.8% Pass@1 accuracy and outperforming state-of-the-art baselines including Alita (75.2%) and Memento (70.9%). Our work establishes a foundation for building truly adaptive agents that continuously evolve their capabilities through principled integration of tool creation and experience accumulation.
LiDAR-GS++:Improving LiDAR Gaussian Reconstruction via Diffusion Priors
Nov 15, 2025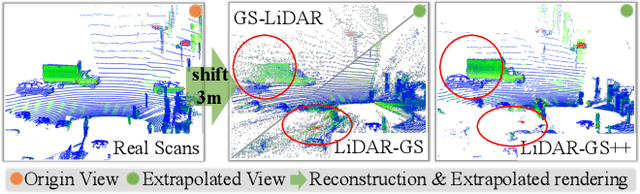
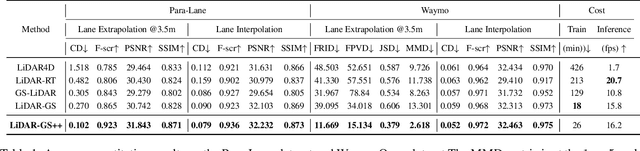
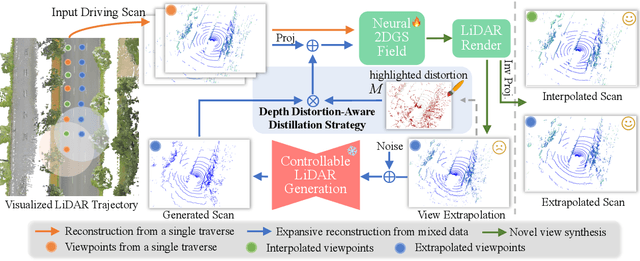
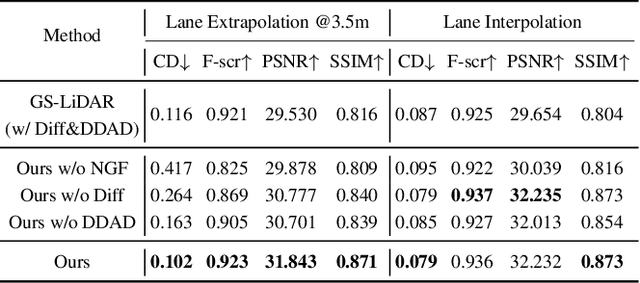
Abstract:Recent GS-based rendering has made significant progress for LiDAR, surpassing Neural Radiance Fields (NeRF) in both quality and speed. However, these methods exhibit artifacts in extrapolated novel view synthesis due to the incomplete reconstruction from single traversal scans. To address this limitation, we present LiDAR-GS++, a LiDAR Gaussian Splatting reconstruction method enhanced by diffusion priors for real-time and high-fidelity re-simulation on public urban roads. Specifically, we introduce a controllable LiDAR generation model conditioned on coarsely extrapolated rendering to produce extra geometry-consistent scans and employ an effective distillation mechanism for expansive reconstruction. By extending reconstruction to under-fitted regions, our approach ensures global geometric consistency for extrapolative novel views while preserving detailed scene surfaces captured by sensors. Experiments on multiple public datasets demonstrate that LiDAR-GS++ achieves state-of-the-art performance for both interpolated and extrapolated viewpoints, surpassing existing GS and NeRF-based methods.
JoyAgent-JDGenie: Technical Report on the GAIA
Oct 01, 2025Abstract:Large Language Models are increasingly deployed as autonomous agents for complex real-world tasks, yet existing systems often focus on isolated improvements without a unifying design for robustness and adaptability. We propose a generalist agent architecture that integrates three core components: a collective multi-agent framework combining planning and execution agents with critic model voting, a hierarchical memory system spanning working, semantic, and procedural layers, and a refined tool suite for search, code execution, and multimodal parsing. Evaluated on a comprehensive benchmark, our framework consistently outperforms open-source baselines and approaches the performance of proprietary systems. These results demonstrate the importance of system-level integration and highlight a path toward scalable, resilient, and adaptive AI assistants capable of operating across diverse domains and tasks.
Cognitive Duality for Adaptive Web Agents
Aug 07, 2025Abstract:Web navigation represents a critical and challenging domain for evaluating artificial general intelligence (AGI), demanding complex decision-making within high-entropy, dynamic environments with combinatorially explosive action spaces. Current approaches to building autonomous web agents either focus on offline imitation learning or online exploration, but rarely integrate both paradigms effectively. Inspired by the dual-process theory of human cognition, we derive a principled decomposition into fast System 1 and slow System 2 cognitive processes. This decomposition provides a unifying perspective on existing web agent methodologies, bridging the gap between offline learning of intuitive reactive behaviors and online acquisition of deliberative planning capabilities. We implement this framework in CogniWeb, a modular agent architecture that adaptively toggles between fast intuitive processing and deliberate reasoning based on task complexity. Our evaluation on WebArena demonstrates that CogniWeb achieves competitive performance (43.96% success rate) while maintaining significantly higher efficiency (75% reduction in token usage).
A Diffusion-Driven Temporal Super-Resolution and Spatial Consistency Enhancement Framework for 4D MRI imaging
Jun 09, 2025Abstract:In medical imaging, 4D MRI enables dynamic 3D visualization, yet the trade-off between spatial and temporal resolution requires prolonged scan time that can compromise temporal fidelity--especially during rapid, large-amplitude motion. Traditional approaches typically rely on registration-based interpolation to generate intermediate frames. However, these methods struggle with large deformations, resulting in misregistration, artifacts, and diminished spatial consistency. To address these challenges, we propose TSSC-Net, a novel framework that generates intermediate frames while preserving spatial consistency. To improve temporal fidelity under fast motion, our diffusion-based temporal super-resolution network generates intermediate frames using the start and end frames as key references, achieving 6x temporal super-resolution in a single inference step. Additionally, we introduce a novel tri-directional Mamba-based module that leverages long-range contextual information to effectively resolve spatial inconsistencies arising from cross-slice misalignment, thereby enhancing volumetric coherence and correcting cross-slice errors. Extensive experiments were performed on the public ACDC cardiac MRI dataset and a real-world dynamic 4D knee joint dataset. The results demonstrate that TSSC-Net can generate high-resolution dynamic MRI from fast-motion data while preserving structural fidelity and spatial consistency.
BioVFM-21M: Benchmarking and Scaling Self-Supervised Vision Foundation Models for Biomedical Image Analysis
May 14, 2025Abstract:Scaling up model and data size have demonstrated impressive performance improvement over a wide range of tasks. Despite extensive studies on scaling behaviors for general-purpose tasks, medical images exhibit substantial differences from natural data. It remains unclear the key factors in developing medical vision foundation models at scale due to the absence of an extensive understanding of scaling behavior in the medical domain. In this paper, we explored the scaling behavior across model sizes, training algorithms, data sizes, and imaging modalities in developing scalable medical vision foundation models by self-supervised learning. To support scalable pretraining, we introduce BioVFM-21M, a large-scale biomedical image dataset encompassing a wide range of biomedical image modalities and anatomies. We observed that scaling up does provide benefits but varies across tasks. Additional analysis reveals several factors correlated with scaling benefits. Finally, we propose BioVFM, a large-scale medical vision foundation model pretrained on 21 million biomedical images, which outperforms the previous state-of-the-art foundation models across 12 medical benchmarks. Our results highlight that while scaling up is beneficial for pursuing better performance, task characteristics, data diversity, pretraining methods, and computational efficiency remain critical considerations for developing scalable medical foundation models.
Industrial-Grade Sensor Simulation via Gaussian Splatting: A Modular Framework for Scalable Editing and Full-Stack Validation
Mar 14, 2025Abstract:Sensor simulation is pivotal for scalable validation of autonomous driving systems, yet existing Neural Radiance Fields (NeRF) based methods face applicability and efficiency challenges in industrial workflows. This paper introduces a Gaussian Splatting (GS) based system to address these challenges: We first break down sensor simulator components and analyze the possible advantages of GS over NeRF. Then in practice, we refactor three crucial components through GS, to leverage its explicit scene representation and real-time rendering: (1) choosing the 2D neural Gaussian representation for physics-compliant scene and sensor modeling, (2) proposing a scene editing pipeline to leverage Gaussian primitives library for data augmentation, and (3) coupling a controllable diffusion model for scene expansion and harmonization. We implement this framework on a proprietary autonomous driving dataset supporting cameras and LiDAR sensors. We demonstrate through ablation studies that our approach reduces frame-wise simulation latency, achieves better geometric and photometric consistency, and enables interpretable explicit scene editing and expansion. Furthermore, we showcase how integrating such a GS-based sensor simulator with traffic and dynamic simulators enables full-stack testing of end-to-end autonomy algorithms. Our work provides both algorithmic insights and practical validation, establishing GS as a cornerstone for industrial-grade sensor simulation.
WEPO: Web Element Preference Optimization for LLM-based Web Navigation
Dec 14, 2024



Abstract:The rapid advancement of autonomous web navigation has significantly benefited from grounding pretrained Large Language Models (LLMs) as agents. However, current research has yet to fully leverage the redundancy of HTML elements for contrastive training. This paper introduces a novel approach to LLM-based web navigation tasks, called Web Element Preference Optimization (WEPO). WEPO utilizes unsupervised preference learning by sampling distance-based non-salient web elements as negative samples, optimizing maximum likelihood objective within Direct Preference Optimization (DPO). We evaluate WEPO on the Mind2Web benchmark and empirically demonstrate that WEPO aligns user high-level intent with output actions more effectively. The results show that our method achieved the state-of-the-art, with an improvement of 13.8% over WebAgent and 5.3% over the visual language model CogAgent baseline. Our findings underscore the potential of preference optimization to enhance web navigation and other web page based tasks, suggesting a promising direction for future research.
Optimized Vessel Segmentation: A Structure-Agnostic Approach with Small Vessel Enhancement and Morphological Correction
Nov 22, 2024Abstract:Accurate segmentation of blood vessels is essential for various clinical assessments and postoperative analyses. However, the inherent challenges of vascular imaging, such as sparsity, fine granularity, low contrast, data distribution variability, and the critical need for preserving topological structure, making generalized vessel segmentation particularly complex. While specialized segmentation methods have been developed for specific anatomical regions, their over-reliance on tailored models hinders broader applicability and generalization. General-purpose segmentation models introduced in medical imaging often fail to address critical vascular characteristics, including the connectivity of segmentation results. To overcome these limitations, we propose an optimized vessel segmentation framework: a structure-agnostic approach incorporating small vessel enhancement and morphological correction for multi-modality vessel segmentation. To train and validate this framework, we compiled a comprehensive multi-modality dataset spanning 17 datasets and benchmarked our model against six SAM-based methods and 17 expert models. The results demonstrate that our approach achieves superior segmentation accuracy, generalization, and a 34.6% improvement in connectivity, underscoring its clinical potential. An ablation study further validates the effectiveness of the proposed improvements. We will release the code and dataset at github following the publication of this work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge