Francesco Galati
Multi-Domain Brain Vessel Segmentation Through Feature Disentanglement
Oct 02, 2025


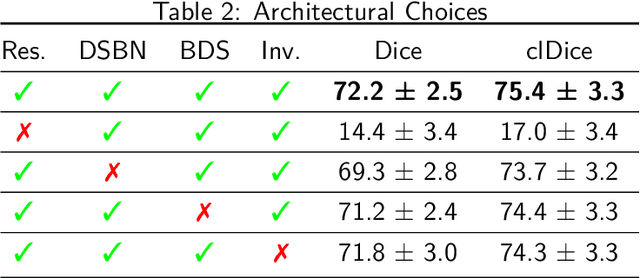
Abstract:The intricate morphology of brain vessels poses significant challenges for automatic segmentation models, which usually focus on a single imaging modality. However, accurately treating brain-related conditions requires a comprehensive understanding of the cerebrovascular tree, regardless of the specific acquisition procedure. Our framework effectively segments brain arteries and veins in various datasets through image-to-image translation while avoiding domain-specific model design and data harmonization between the source and the target domain. This is accomplished by employing disentanglement techniques to independently manipulate different image properties, allowing them to move from one domain to another in a label-preserving manner. Specifically, we focus on manipulating vessel appearances during adaptation while preserving spatial information, such as shapes and locations, which are crucial for correct segmentation. Our evaluation effectively bridges large and varied domain gaps across medical centers, image modalities, and vessel types. Additionally, we conduct ablation studies on the optimal number of required annotations and other architectural choices. The results highlight our framework's robustness and versatility, demonstrating the potential of domain adaptation methodologies to perform cerebrovascular image segmentation in multiple scenarios accurately. Our code is available at https://github.com/i-vesseg/MultiVesSeg.
* 19 pages, 7 figures, 3 tables. Joint first authors: Francesco Galati and Daniele Falcetta. Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2025:021. Code available at https://github.com/i-vesseg/MultiVesSeg
ImageSet2Text: Describing Sets of Images through Text
Mar 25, 2025



Abstract:We introduce ImageSet2Text, a novel approach that leverages vision-language foundation models to automatically create natural language descriptions of image sets. Inspired by concept bottleneck models (CBMs) and based on visual-question answering (VQA) chains, ImageSet2Text iteratively extracts key concepts from image subsets, encodes them into a structured graph, and refines insights using an external knowledge graph and CLIP-based validation. This iterative process enhances interpretability and enables accurate and detailed set-level summarization. Through extensive experiments, we evaluate ImageSet2Text's descriptions on accuracy, completeness, readability and overall quality, benchmarking it against existing vision-language models and introducing new datasets for large-scale group image captioning.
HyperMM : Robust Multimodal Learning with Varying-sized Inputs
Jul 30, 2024



Abstract:Combining multiple modalities carrying complementary information through multimodal learning (MML) has shown considerable benefits for diagnosing multiple pathologies. However, the robustness of multimodal models to missing modalities is often overlooked. Most works assume modality completeness in the input data, while in clinical practice, it is common to have incomplete modalities. Existing solutions that address this issue rely on modality imputation strategies before using supervised learning models. These strategies, however, are complex, computationally costly and can strongly impact subsequent prediction models. Hence, they should be used with parsimony in sensitive applications such as healthcare. We propose HyperMM, an end-to-end framework designed for learning with varying-sized inputs. Specifically, we focus on the task of supervised MML with missing imaging modalities without using imputation before training. We introduce a novel strategy for training a universal feature extractor using a conditional hypernetwork, and propose a permutation-invariant neural network that can handle inputs of varying dimensions to process the extracted features, in a two-phase task-agnostic framework. We experimentally demonstrate the advantages of our method in two tasks: Alzheimer's disease detection and breast cancer classification. We demonstrate that our strategy is robust to high rates of missing data and that its flexibility allows it to handle varying-sized datasets beyond the scenario of missing modalities.
Benchmarking the CoW with the TopCoW Challenge: Topology-Aware Anatomical Segmentation of the Circle of Willis for CTA and MRA
Dec 29, 2023



Abstract:The Circle of Willis (CoW) is an important network of arteries connecting major circulations of the brain. Its vascular architecture is believed to affect the risk, severity, and clinical outcome of serious neuro-vascular diseases. However, characterizing the highly variable CoW anatomy is still a manual and time-consuming expert task. The CoW is usually imaged by two angiographic imaging modalities, magnetic resonance angiography (MRA) and computed tomography angiography (CTA), but there exist limited public datasets with annotations on CoW anatomy, especially for CTA. Therefore we organized the TopCoW Challenge in 2023 with the release of an annotated CoW dataset and invited submissions worldwide for the CoW segmentation task, which attracted over 140 registered participants from four continents. TopCoW dataset was the first public dataset with voxel-level annotations for CoW's 13 vessel components, made possible by virtual-reality (VR) technology. It was also the first dataset with paired MRA and CTA from the same patients. TopCoW challenge aimed to tackle the CoW characterization problem as a multiclass anatomical segmentation task with an emphasis on topological metrics. The top performing teams managed to segment many CoW components to Dice scores around 90%, but with lower scores for communicating arteries and rare variants. There were also topological mistakes for predictions with high Dice scores. Additional topological analysis revealed further areas for improvement in detecting certain CoW components and matching CoW variant's topology accurately. TopCoW represented a first attempt at benchmarking the CoW anatomical segmentation task for MRA and CTA, both morphologically and topologically.
A2V: A Semi-Supervised Domain Adaptation Framework for Brain Vessel Segmentation via Two-Phase Training Angiography-to-Venography Translation
Sep 12, 2023



Abstract:We present a semi-supervised domain adaptation framework for brain vessel segmentation from different image modalities. Existing state-of-the-art methods focus on a single modality, despite the wide range of available cerebrovascular imaging techniques. This can lead to significant distribution shifts that negatively impact the generalization across modalities. By relying on annotated angiographies and a limited number of annotated venographies, our framework accomplishes image-to-image translation and semantic segmentation, leveraging a disentangled and semantically rich latent space to represent heterogeneous data and perform image-level adaptation from source to target domains. Moreover, we reduce the typical complexity of cycle-based architectures and minimize the use of adversarial training, which allows us to build an efficient and intuitive model with stable training. We evaluate our method on magnetic resonance angiographies and venographies. While achieving state-of-the-art performance in the source domain, our method attains a Dice score coefficient in the target domain that is only 8.9% lower, highlighting its promising potential for robust cerebrovascular image segmentation across different modalities.
Binary domain generalization for sparsifying binary neural networks
Jun 23, 2023Abstract:Binary neural networks (BNNs) are an attractive solution for developing and deploying deep neural network (DNN)-based applications in resource constrained devices. Despite their success, BNNs still suffer from a fixed and limited compression factor that may be explained by the fact that existing pruning methods for full-precision DNNs cannot be directly applied to BNNs. In fact, weight pruning of BNNs leads to performance degradation, which suggests that the standard binarization domain of BNNs is not well adapted for the task. This work proposes a novel more general binary domain that extends the standard binary one that is more robust to pruning techniques, thus guaranteeing improved compression and avoiding severe performance losses. We demonstrate a closed-form solution for quantizing the weights of a full-precision network into the proposed binary domain. Finally, we show the flexibility of our method, which can be combined with other pruning strategies. Experiments over CIFAR-10 and CIFAR-100 demonstrate that the novel approach is able to generate efficient sparse networks with reduced memory usage and run-time latency, while maintaining performance.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Efficient Model Monitoring for Quality Control in Cardiac Image Segmentation
Apr 12, 2021



Abstract:Deep learning methods have reached state-of-the-art performance in cardiac image segmentation. Currently, the main bottleneck towards their effective translation into clinics requires assuring continuous high model performance and segmentation results. In this work, we present a novel learning framework to monitor the performance of heart segmentation models in the absence of ground truth. Formulated as an anomaly detection problem, the monitoring framework allows deriving surrogate quality measures for a segmentation and allows flagging suspicious results. We propose two different types of quality measures, a global score and a pixel-wise map. We demonstrate their use by reproducing the final rankings of a cardiac segmentation challenge in the absence of ground truth. Results show that our framework is accurate, fast, and scalable, confirming it is a viable option for quality control monitoring in clinical practice and large population studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge