Fandong Zhang
LDRNet: Large Deformation Registration Model for Chest CT Registration
Feb 02, 2026Abstract:Most of the deep learning based medical image registration algorithms focus on brain image registration tasks.Compared with brain registration, the chest CT registration has larger deformation, more complex background and region over-lap. In this paper, we propose a fast unsupervised deep learning method, LDRNet, for large deformation image registration of chest CT images. We first predict a coarse resolution registration field, then refine it from coarse to fine. We propose two innovative technical components: 1) a refine block that is used to refine the registration field in different resolutions, 2) a rigid block that is used to learn transformation matrix from high-level features. We train and evaluate our model on the private dataset and public dataset SegTHOR. We compare our performance with state-of-the-art traditional registration methods as well as deep learning registration models VoxelMorph, RCN, and LapIRN. The results demonstrate that our model achieves state-of-the-art performance for large deformation images registration and is much faster.
Autoregressive Sequence Modeling for 3D Medical Image Representation
Sep 13, 2024



Abstract:Three-dimensional (3D) medical images, such as Computed Tomography (CT) and Magnetic Resonance Imaging (MRI), are essential for clinical applications. However, the need for diverse and comprehensive representations is particularly pronounced when considering the variability across different organs, diagnostic tasks, and imaging modalities. How to effectively interpret the intricate contextual information and extract meaningful insights from these images remains an open challenge to the community. While current self-supervised learning methods have shown potential, they often consider an image as a whole thereby overlooking the extensive, complex relationships among local regions from one or multiple images. In this work, we introduce a pioneering method for learning 3D medical image representations through an autoregressive pre-training framework. Our approach sequences various 3D medical images based on spatial, contrast, and semantic correlations, treating them as interconnected visual tokens within a token sequence. By employing an autoregressive sequence modeling task, we predict the next visual token in the sequence, which allows our model to deeply understand and integrate the contextual information inherent in 3D medical images. Additionally, we implement a random startup strategy to avoid overestimating token relationships and to enhance the robustness of learning. The effectiveness of our approach is demonstrated by the superior performance over others on nine downstream tasks in public datasets.
Cross-Dimensional Medical Self-Supervised Representation Learning Based on a Pseudo-3D Transformation
Jun 03, 2024



Abstract:Medical image analysis suffers from a shortage of data, whether annotated or not. This becomes even more pronounced when it comes to 3D medical images. Self-Supervised Learning (SSL) can partially ease this situation by using unlabeled data. However, most existing SSL methods can only make use of data in a single dimensionality (e.g. 2D or 3D), and are incapable of enlarging the training dataset by using data with differing dimensionalities jointly. In this paper, we propose a new cross-dimensional SSL framework based on a pseudo-3D transformation (CDSSL-P3D), that can leverage both 2D and 3D data for joint pre-training. Specifically, we introduce an image transformation based on the im2col algorithm, which converts 2D images into a format consistent with 3D data. This transformation enables seamless integration of 2D and 3D data, and facilitates cross-dimensional self-supervised learning for 3D medical image analysis. We run extensive experiments on 13 downstream tasks, including 2D and 3D classification and segmentation. The results indicate that our CDSSL-P3D achieves superior performance, outperforming other advanced SSL methods.
Graph Convolution Based Cross-Network Multi-Scale Feature Fusion for Deep Vessel Segmentation
Jan 06, 2023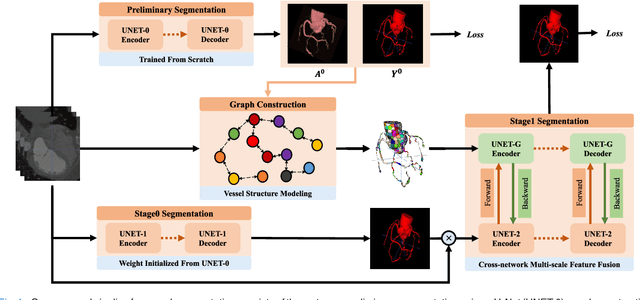
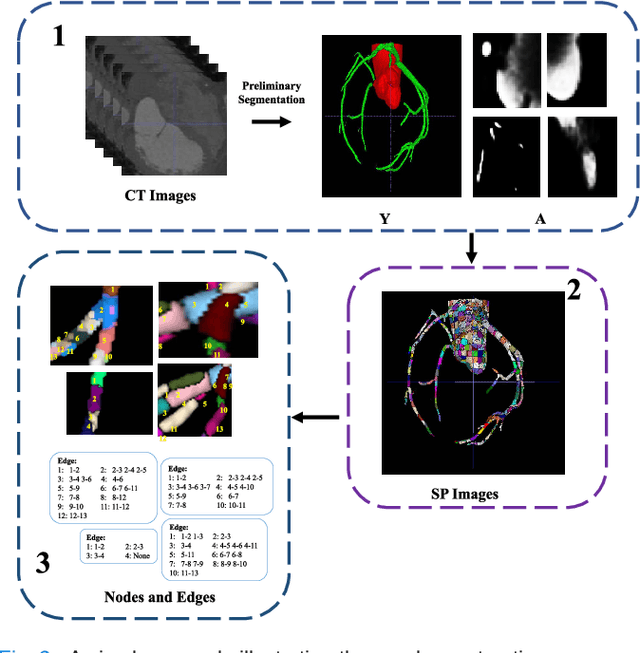
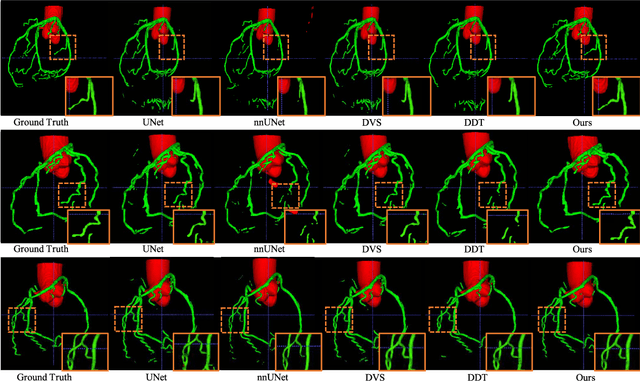
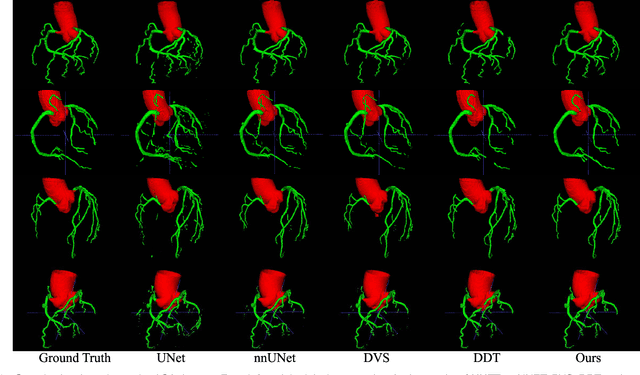
Abstract:Vessel segmentation is widely used to help with vascular disease diagnosis. Vessels reconstructed using existing methods are often not sufficiently accurate to meet clinical use standards. This is because 3D vessel structures are highly complicated and exhibit unique characteristics, including sparsity and anisotropy. In this paper, we propose a novel hybrid deep neural network for vessel segmentation. Our network consists of two cascaded subnetworks performing initial and refined segmentation respectively. The second subnetwork further has two tightly coupled components, a traditional CNN-based U-Net and a graph U-Net. Cross-network multi-scale feature fusion is performed between these two U-shaped networks to effectively support high-quality vessel segmentation. The entire cascaded network can be trained from end to end. The graph in the second subnetwork is constructed according to a vessel probability map as well as appearance and semantic similarities in the original CT volume. To tackle the challenges caused by the sparsity and anisotropy of vessels, a higher percentage of graph nodes are distributed in areas that potentially contain vessels while a higher percentage of edges follow the orientation of potential nearbyvessels. Extensive experiments demonstrate our deep network achieves state-of-the-art 3D vessel segmentation performance on multiple public and in-house datasets.
Domain Invariant Model with Graph Convolutional Network for Mammogram Classification
Apr 21, 2022



Abstract:Due to its safety-critical property, the image-based diagnosis is desired to achieve robustness on out-of-distribution (OOD) samples. A natural way towards this goal is capturing only clinically disease-related features, which is composed of macroscopic attributes (e.g., margins, shapes) and microscopic image-based features (e.g., textures) of lesion-related areas. However, such disease-related features are often interweaved with data-dependent (but disease irrelevant) biases during learning, disabling the OOD generalization. To resolve this problem, we propose a novel framework, namely Domain Invariant Model with Graph Convolutional Network (DIM-GCN), which only exploits invariant disease-related features from multiple domains. Specifically, we first propose a Bayesian network, which explicitly decomposes the latent variables into disease-related and other disease-irrelevant parts that are provable to be disentangled from each other. Guided by this, we reformulate the objective function based on Variational Auto-Encoder, in which the encoder in each domain has two branches: the domain-independent and -dependent ones, which respectively encode disease-related and -irrelevant features. To better capture the macroscopic features, we leverage the observed clinical attributes as a goal for reconstruction, via Graph Convolutional Network (GCN). Finally, we only implement the disease-related features for prediction. The effectiveness and utility of our method are demonstrated by the superior OOD generalization performance over others on mammogram benign/malignant diagnosis.
Context-LGM: Leveraging Object-Context Relation for Context-Aware Object Recognition
Oct 08, 2021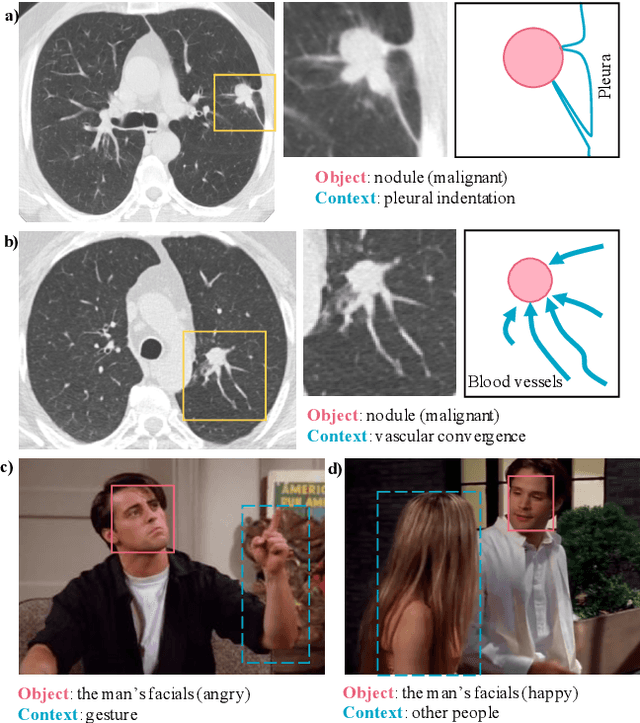
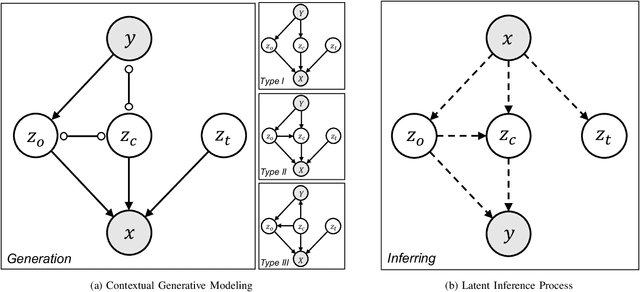
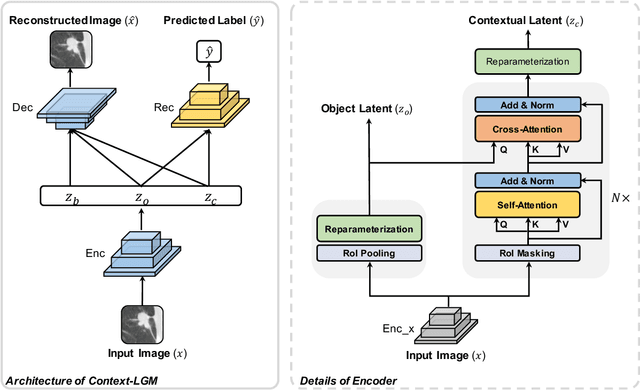
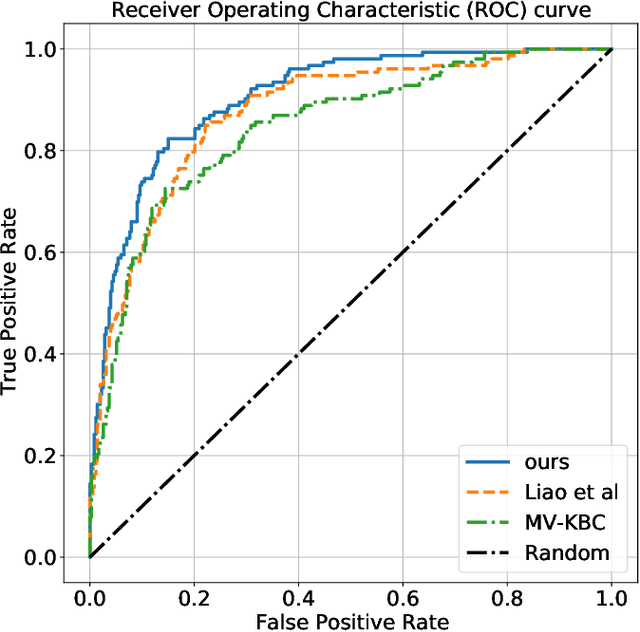
Abstract:Context, as referred to situational factors related to the object of interest, can help infer the object's states or properties in visual recognition. As such contextual features are too diverse (across instances) to be annotated, existing attempts simply exploit image labels as supervision to learn them, resulting in various contextual tricks, such as features pyramid, context attention, etc. However, without carefully modeling the context's properties, especially its relation to the object, their estimated context can suffer from large inaccuracy. To amend this problem, we propose a novel Contextual Latent Generative Model (Context-LGM), which considers the object-context relation and models it in a hierarchical manner. Specifically, we firstly introduce a latent generative model with a pair of correlated latent variables to respectively model the object and context, and embed their correlation via the generative process. Then, to infer contextual features, we reformulate the objective function of Variational Auto-Encoder (VAE), where contextual features are learned as a posterior distribution conditioned on the object. Finally, to implement this contextual posterior, we introduce a Transformer that takes the object's information as a reference and locates correlated contextual factors. The effectiveness of our method is verified by state-of-the-art performance on two context-aware object recognition tasks, i.e. lung cancer prediction and emotion recognition.
Act Like a Radiologist: Towards Reliable Multi-view Correspondence Reasoning for Mammogram Mass Detection
May 21, 2021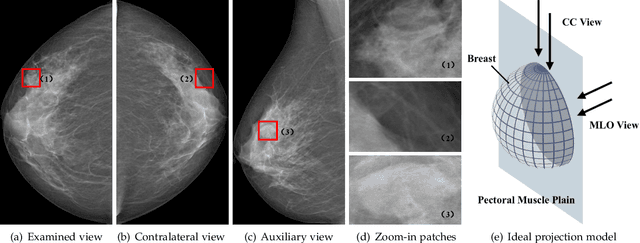
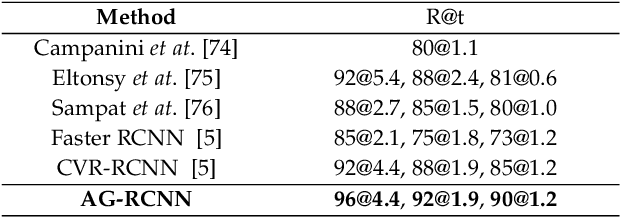
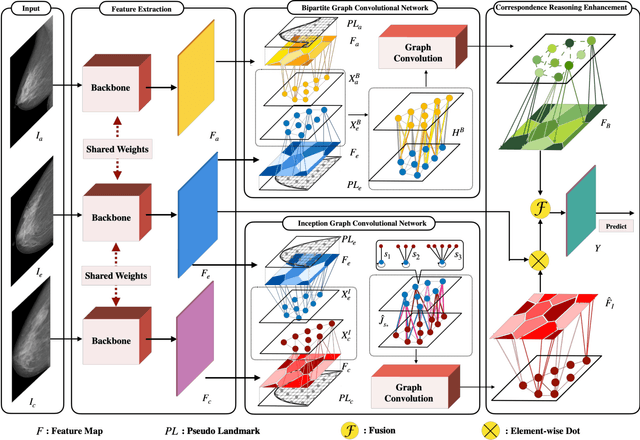
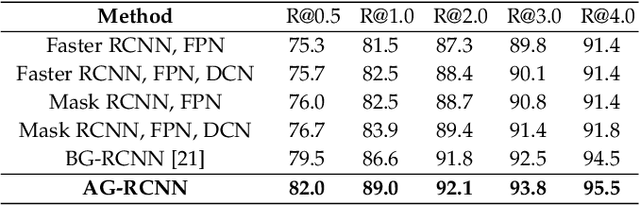
Abstract:Mammogram mass detection is crucial for diagnosing and preventing the breast cancers in clinical practice. The complementary effect of multi-view mammogram images provides valuable information about the breast anatomical prior structure and is of great significance in digital mammography interpretation. However, unlike radiologists who can utilize the natural reasoning ability to identify masses based on multiple mammographic views, how to endow the existing object detection models with the capability of multi-view reasoning is vital for decision-making in clinical diagnosis but remains the boundary to explore. In this paper, we propose an Anatomy-aware Graph convolutional Network (AGN), which is tailored for mammogram mass detection and endows existing detection methods with multi-view reasoning ability. The proposed AGN consists of three steps. Firstly, we introduce a Bipartite Graph convolutional Network (BGN) to model the intrinsic geometric and semantic relations of ipsilateral views. Secondly, considering that the visual asymmetry of bilateral views is widely adopted in clinical practice to assist the diagnosis of breast lesions, we propose an Inception Graph convolutional Network (IGN) to model the structural similarities of bilateral views. Finally, based on the constructed graphs, the multi-view information is propagated through nodes methodically, which equips the features learned from the examined view with multi-view reasoning ability. Experiments on two standard benchmarks reveal that AGN significantly exceeds the state-of-the-art performance. Visualization results show that AGN provides interpretable visual cues for clinical diagnosis.
Bilateral Asymmetry Guided Counterfactual Generating Network for Mammogram Classification
Sep 30, 2020



Abstract:Mammogram benign or malignant classification with only image-level labels is challenging due to the absence of lesion annotations. Motivated by the symmetric prior that the lesions on one side of breasts rarely appear in the corresponding areas on the other side, given a diseased image, we can explore a counterfactual problem that how would the features have behaved if there were no lesions in the image, so as to identify the lesion areas. We derive a new theoretical result for counterfactual generation based on the symmetric prior. By building a causal model that entails such a prior for bilateral images, we obtain two optimization goals for counterfactual generation, which can be accomplished via our newly proposed counterfactual generative network. Our proposed model is mainly composed of Generator Adversarial Network and a \emph{prediction feedback mechanism}, they are optimized jointly and prompt each other. Specifically, the former can further improve the classification performance by generating counterfactual features to calculate lesion areas. On the other hand, the latter helps counterfactual generation by the supervision of classification loss. The utility of our method and the effectiveness of each module in our model can be verified by state-of-the-art performance on INBreast and an in-house dataset and ablation studies.
Integrating Feature and Image Pyramid: A Lung Nodule Detector Learned in Curriculum Fashion
Aug 01, 2018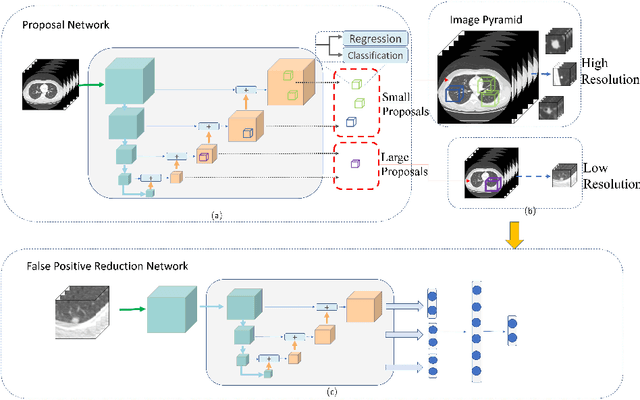
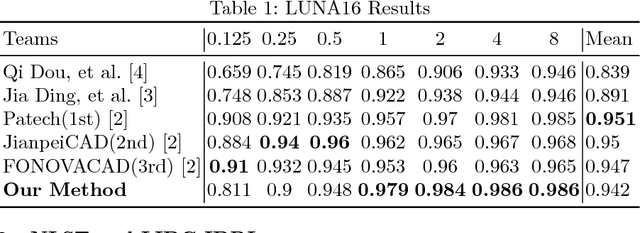
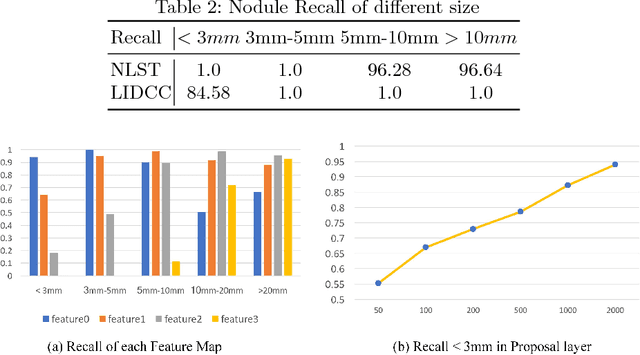

Abstract:Lung nodules suffer large variation in size and appearance in CT images. Nodules less than 10mm can easily lose information after down-sampling in convolutional neural networks, which results in low sensitivity. In this paper, a combination of 3D image and feature pyramid is exploited to integrate lower-level texture features with high-level semantic features, thus leading to a higher recall. However, 3D operations are time and memory consuming, which aggravates the situation with the explosive growth of medical images. To tackle this problem, we propose a general curriculum training strategy to speed up training. An dynamic sampling method is designed to pick up partial samples which give the best contribution to network training, thus leading to much less time consuming. In experiments, we demonstrate that the proposed network outperforms previous state-of-the-art methods. Meanwhile, our sampling strategy halves the training time of the proposal network on LUNA16.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge