Churan Wang
VLM Can Be a Good Assistant: Enhancing Embodied Visual Tracking with Self-Improving Vision-Language Models
May 28, 2025Abstract:We introduce a novel self-improving framework that enhances Embodied Visual Tracking (EVT) with Vision-Language Models (VLMs) to address the limitations of current active visual tracking systems in recovering from tracking failure. Our approach combines the off-the-shelf active tracking methods with VLMs' reasoning capabilities, deploying a fast visual policy for normal tracking and activating VLM reasoning only upon failure detection. The framework features a memory-augmented self-reflection mechanism that enables the VLM to progressively improve by learning from past experiences, effectively addressing VLMs' limitations in 3D spatial reasoning. Experimental results demonstrate significant performance improvements, with our framework boosting success rates by $72\%$ with state-of-the-art RL-based approaches and $220\%$ with PID-based methods in challenging environments. This work represents the first integration of VLM-based reasoning to assist EVT agents in proactive failure recovery, offering substantial advances for real-world robotic applications that require continuous target monitoring in dynamic, unstructured environments. Project website: https://sites.google.com/view/evt-recovery-assistant.
Hierarchical Instruction-aware Embodied Visual Tracking
May 27, 2025Abstract:User-Centric Embodied Visual Tracking (UC-EVT) presents a novel challenge for reinforcement learning-based models due to the substantial gap between high-level user instructions and low-level agent actions. While recent advancements in language models (e.g., LLMs, VLMs, VLAs) have improved instruction comprehension, these models face critical limitations in either inference speed (LLMs, VLMs) or generalizability (VLAs) for UC-EVT tasks. To address these challenges, we propose \textbf{Hierarchical Instruction-aware Embodied Visual Tracking (HIEVT)} agent, which bridges instruction comprehension and action generation using \textit{spatial goals} as intermediaries. HIEVT first introduces \textit{LLM-based Semantic-Spatial Goal Aligner} to translate diverse human instructions into spatial goals that directly annotate the desired spatial position. Then the \textit{RL-based Adaptive Goal-Aligned Policy}, a general offline policy, enables the tracker to position the target as specified by the spatial goal. To benchmark UC-EVT tasks, we collect over ten million trajectories for training and evaluate across one seen environment and nine unseen challenging environments. Extensive experiments and real-world deployments demonstrate the robustness and generalizability of HIEVT across diverse environments, varying target dynamics, and complex instruction combinations. The complete project is available at https://sites.google.com/view/hievt.
Clinical Inspired MRI Lesion Segmentation
Feb 22, 2025Abstract:Magnetic resonance imaging (MRI) is a potent diagnostic tool for detecting pathological tissues in various diseases. Different MRI sequences have different contrast mechanisms and sensitivities for different types of lesions, which pose challenges to accurate and consistent lesion segmentation. In clinical practice, radiologists commonly use the sub-sequence feature, i.e. the difference between post contrast-enhanced T1-weighted (post) and pre-contrast-enhanced (pre) sequences, to locate lesions. Inspired by this, we propose a residual fusion method to learn subsequence representation for MRI lesion segmentation. Specifically, we iteratively and adaptively fuse features from pre- and post-contrast sequences at multiple resolutions, using dynamic weights to achieve optimal fusion and address diverse lesion enhancement patterns. Our method achieves state-of-the-art performances on BraTS2023 dataset for brain tumor segmentation and our in-house breast MRI dataset for breast lesion segmentation. Our method is clinically inspired and has the potential to facilitate lesion segmentation in various applications.
UnrealZoo: Enriching Photo-realistic Virtual Worlds for Embodied AI
Dec 30, 2024



Abstract:We introduce UnrealZoo, a rich collection of photo-realistic 3D virtual worlds built on Unreal Engine, designed to reflect the complexity and variability of the open worlds. Additionally, we offer a variety of playable entities for embodied AI agents. Based on UnrealCV, we provide a suite of easy-to-use Python APIs and tools for various potential applications, such as data collection, environment augmentation, distributed training, and benchmarking. We optimize the rendering and communication efficiency of UnrealCV to support advanced applications, such as multi-agent interaction. Our experiments benchmark agents in various complex scenes, focusing on visual navigation and tracking, which are fundamental capabilities for embodied visual intelligence. The results yield valuable insights into the advantages of diverse training environments for reinforcement learning (RL) agents and the challenges faced by current embodied vision agents, including those based on RL and large vision-language models (VLMs), in open worlds. These challenges involve latency in closed-loop control in dynamic scenes and reasoning about 3D spatial structures in unstructured terrain.
Enhanced MRI Representation via Cross-series Masking
Dec 10, 2024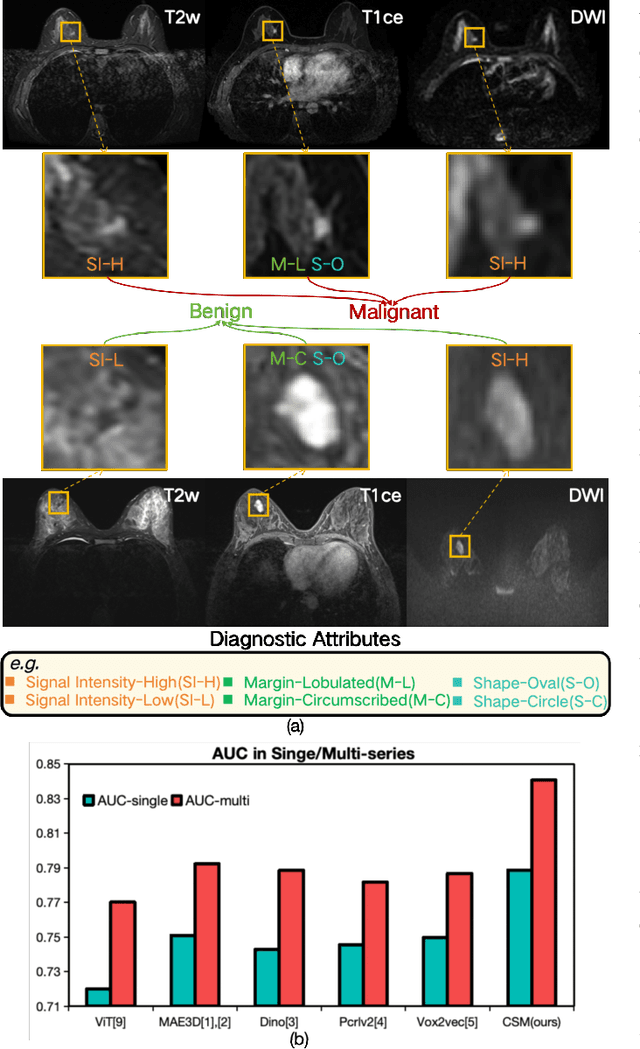
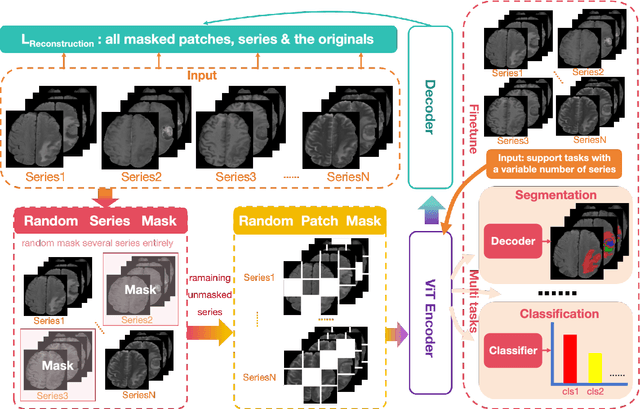
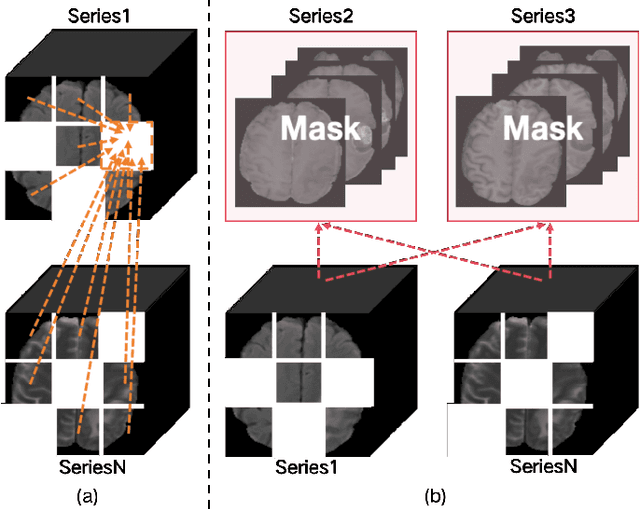
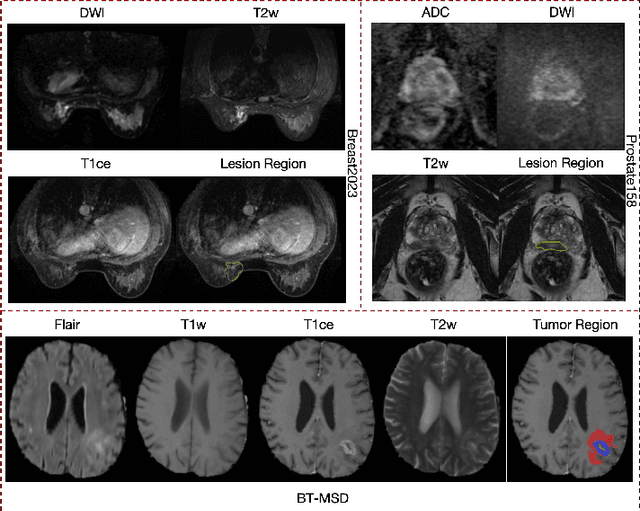
Abstract:Magnetic resonance imaging (MRI) is indispensable for diagnosing and planning treatment in various medical conditions due to its ability to produce multi-series images that reveal different tissue characteristics. However, integrating these diverse series to form a coherent analysis presents significant challenges, such as differing spatial resolutions and contrast patterns meanwhile requiring extensive annotated data, which is scarce in clinical practice. Due to these issues, we introduce a novel Cross-Series Masking (CSM) Strategy for effectively learning MRI representation in a self-supervised manner. Specifically, CSM commences by randomly sampling a subset of regions and series, which are then strategically masked. In the training process, the cross-series representation is learned by utilizing the unmasked data to reconstruct the masked portions. This process not only integrates information across different series but also facilitates the ability to model both intra-series and inter-series correlations and complementarities. With the learned representation, the downstream tasks like segmentation and classification are also enhanced. Taking brain tissue segmentation, breast tumor benign/malignant classification, and prostate cancer diagnosis as examples, our method achieves state-of-the-art performance on both public and in-house datasets.
Autoregressive Sequence Modeling for 3D Medical Image Representation
Sep 13, 2024



Abstract:Three-dimensional (3D) medical images, such as Computed Tomography (CT) and Magnetic Resonance Imaging (MRI), are essential for clinical applications. However, the need for diverse and comprehensive representations is particularly pronounced when considering the variability across different organs, diagnostic tasks, and imaging modalities. How to effectively interpret the intricate contextual information and extract meaningful insights from these images remains an open challenge to the community. While current self-supervised learning methods have shown potential, they often consider an image as a whole thereby overlooking the extensive, complex relationships among local regions from one or multiple images. In this work, we introduce a pioneering method for learning 3D medical image representations through an autoregressive pre-training framework. Our approach sequences various 3D medical images based on spatial, contrast, and semantic correlations, treating them as interconnected visual tokens within a token sequence. By employing an autoregressive sequence modeling task, we predict the next visual token in the sequence, which allows our model to deeply understand and integrate the contextual information inherent in 3D medical images. Additionally, we implement a random startup strategy to avoid overestimating token relationships and to enhance the robustness of learning. The effectiveness of our approach is demonstrated by the superior performance over others on nine downstream tasks in public datasets.
Cross-Dimensional Medical Self-Supervised Representation Learning Based on a Pseudo-3D Transformation
Jun 03, 2024



Abstract:Medical image analysis suffers from a shortage of data, whether annotated or not. This becomes even more pronounced when it comes to 3D medical images. Self-Supervised Learning (SSL) can partially ease this situation by using unlabeled data. However, most existing SSL methods can only make use of data in a single dimensionality (e.g. 2D or 3D), and are incapable of enlarging the training dataset by using data with differing dimensionalities jointly. In this paper, we propose a new cross-dimensional SSL framework based on a pseudo-3D transformation (CDSSL-P3D), that can leverage both 2D and 3D data for joint pre-training. Specifically, we introduce an image transformation based on the im2col algorithm, which converts 2D images into a format consistent with 3D data. This transformation enables seamless integration of 2D and 3D data, and facilitates cross-dimensional self-supervised learning for 3D medical image analysis. We run extensive experiments on 13 downstream tasks, including 2D and 3D classification and segmentation. The results indicate that our CDSSL-P3D achieves superior performance, outperforming other advanced SSL methods.
Empowering Embodied Visual Tracking with Visual Foundation Models and Offline RL
Apr 15, 2024



Abstract:Embodied visual tracking is to follow a target object in dynamic 3D environments using an agent's egocentric vision. This is a vital and challenging skill for embodied agents. However, existing methods suffer from inefficient training and poor generalization. In this paper, we propose a novel framework that combines visual foundation models (VFM) and offline reinforcement learning (offline RL) to empower embodied visual tracking. We use a pre-trained VFM, such as ``Tracking Anything", to extract semantic segmentation masks with text prompts. We then train a recurrent policy network with offline RL, e.g., Conservative Q-Learning, to learn from the collected demonstrations without online agent-environment interactions. To further improve the robustness and generalization of the policy network, we also introduce a mask re-targeting mechanism and a multi-level data collection strategy. In this way, we can train a robust tracker within an hour on a consumer-level GPU, e.g., Nvidia RTX 3090. Such efficiency is unprecedented for RL-based visual tracking methods. We evaluate our tracker on several high-fidelity environments with challenging situations, such as distraction and occlusion. The results show that our agent outperforms state-of-the-art methods in terms of sample efficiency, robustness to distractors, and generalization to unseen scenarios and targets. We also demonstrate the transferability of the learned tracker from the virtual world to real-world scenarios.
Semi- and Weakly-Supervised Learning for Mammogram Mass Segmentation with Limited Annotations
Mar 14, 2024



Abstract:Accurate identification of breast masses is crucial in diagnosing breast cancer; however, it can be challenging due to their small size and being camouflaged in surrounding normal glands. Worse still, it is also expensive in clinical practice to obtain adequate pixel-wise annotations for training deep neural networks. To overcome these two difficulties with one stone, we propose a semi- and weakly-supervised learning framework for mass segmentation that utilizes limited strongly-labeled samples and sufficient weakly-labeled samples to achieve satisfactory performance. The framework consists of an auxiliary branch to exclude lesion-irrelevant background areas, a segmentation branch for final prediction, and a spatial prompting module to integrate the complementary information of the two branches. We further disentangle encoded obscure features into lesion-related and others to boost performance. Experiments on CBIS-DDSM and INbreast datasets demonstrate the effectiveness of our method.
Domain Invariant Model with Graph Convolutional Network for Mammogram Classification
Apr 21, 2022



Abstract:Due to its safety-critical property, the image-based diagnosis is desired to achieve robustness on out-of-distribution (OOD) samples. A natural way towards this goal is capturing only clinically disease-related features, which is composed of macroscopic attributes (e.g., margins, shapes) and microscopic image-based features (e.g., textures) of lesion-related areas. However, such disease-related features are often interweaved with data-dependent (but disease irrelevant) biases during learning, disabling the OOD generalization. To resolve this problem, we propose a novel framework, namely Domain Invariant Model with Graph Convolutional Network (DIM-GCN), which only exploits invariant disease-related features from multiple domains. Specifically, we first propose a Bayesian network, which explicitly decomposes the latent variables into disease-related and other disease-irrelevant parts that are provable to be disentangled from each other. Guided by this, we reformulate the objective function based on Variational Auto-Encoder, in which the encoder in each domain has two branches: the domain-independent and -dependent ones, which respectively encode disease-related and -irrelevant features. To better capture the macroscopic features, we leverage the observed clinical attributes as a goal for reconstruction, via Graph Convolutional Network (GCN). Finally, we only implement the disease-related features for prediction. The effectiveness and utility of our method are demonstrated by the superior OOD generalization performance over others on mammogram benign/malignant diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge