Emily Zhao
FD-GATDR: A Federated-Decentralized-Learning Graph Attention Network for Doctor Recommendation Using EHR
Jul 11, 2022



Abstract:In the past decade, with the development of big data technology, an increasing amount of patient information has been stored as electronic health records (EHRs). Leveraging these data, various doctor recommendation systems have been proposed. Typically, such studies process the EHR data in a flat-structured manner, where each encounter was treated as an unordered set of features. Nevertheless, the heterogeneous structured information such as service sequence stored in claims shall not be ignored. This paper presents a doctor recommendation system with time embedding to reconstruct the potential connections between patients and doctors using heterogeneous graph attention network. Besides, to address the privacy issue of patient data sharing crossing hospitals, a federated decentralized learning method based on a minimization optimization model is also proposed. The graph-based recommendation system has been validated on a EHR dataset. Compared to baseline models, the proposed method improves the AUC by up to 6.2%. And our proposed federated-based algorithm not only yields the fictitious fusion center's performance but also enjoys a convergence rate of O(1/T).
eTREE: Learning Tree-structured Embeddings
Dec 20, 2020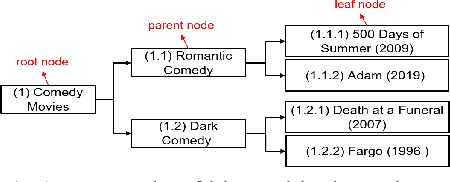
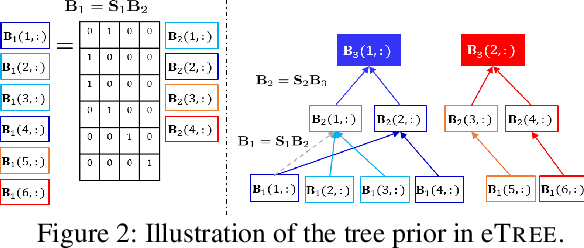
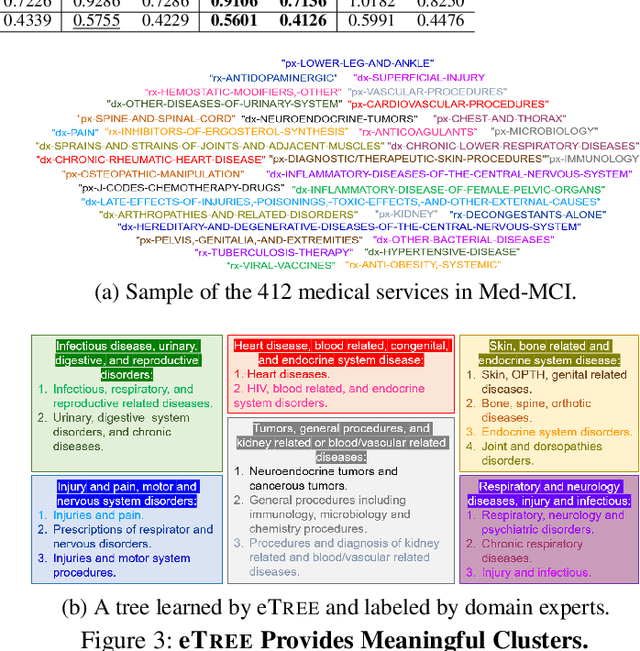
Abstract:Matrix factorization (MF) plays an important role in a wide range of machine learning and data mining models. MF is commonly used to obtain item embeddings and feature representations due to its ability to capture correlations and higher-order statistical dependencies across dimensions. In many applications, the categories of items exhibit a hierarchical tree structure. For instance, human diseases can be divided into coarse categories, e.g., bacterial, and viral. These categories can be further divided into finer categories, e.g., viral infections can be respiratory, gastrointestinal, and exanthematous viral diseases. In e-commerce, products, movies, books, etc., are grouped into hierarchical categories, e.g., clothing items are divided by gender, then by type (formal, casual, etc.). While the tree structure and the categories of the different items may be known in some applications, they have to be learned together with the embeddings in many others. In this work, we propose eTREE, a model that incorporates the (usually ignored) tree structure to enhance the quality of the embeddings. We leverage the special uniqueness properties of Nonnegative MF (NMF) to prove identifiability of eTREE. The proposed model not only exploits the tree structure prior, but also learns the hierarchical clustering in an unsupervised data-driven fashion. We derive an efficient algorithmic solution and a scalable implementation of eTREE that exploits parallel computing, computation caching, and warm start strategies. We showcase the effectiveness of eTREE on real data from various application domains: healthcare, recommender systems, and education. We also demonstrate the meaningfulness of the tree obtained from eTREE by means of domain experts interpretation.
Representation Learning of EHR Data via Graph-Based Medical Entity Embedding
Oct 07, 2019
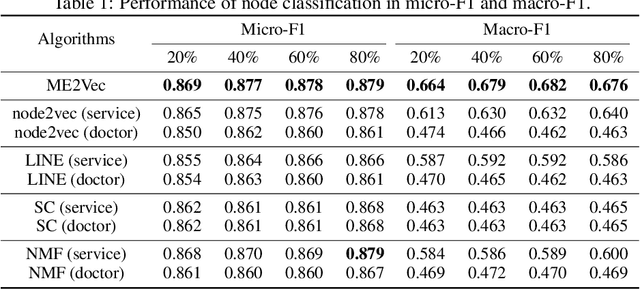
Abstract:Automatic representation learning of key entities in electronic health record (EHR) data is a critical step for healthcare informatics that turns heterogeneous medical records into structured and actionable information. Here we propose ME2Vec, an algorithmic framework for learning low-dimensional vectors of the most common entities in EHR: medical services, doctors, and patients. ME2Vec leverages diverse graph embedding techniques to cater for the unique characteristic of each medical entity. Using real-world clinical data, we demonstrate the efficacy of ME2Vec over competitive baselines on disease diagnosis prediction.
Predicting Treatment Initiation from Clinical Time Series Data via Graph-Augmented Time-Sensitive Model
Jul 01, 2019



Abstract:Many computational models were proposed to extract temporal patterns from clinical time series for each patient and among patient group for predictive healthcare. However, the common relations among patients (e.g., share the same doctor) were rarely considered. In this paper, we represent patients and clinicians relations by bipartite graphs addressing for example from whom a patient get a diagnosis. We then solve for the top eigenvectors of the graph Laplacian, and include the eigenvectors as latent representations of the similarity between patient-clinician pairs into a time-sensitive prediction model. We conducted experiments using real-world data to predict the initiation of first-line treatment for Chronic Lymphocytic Leukemia (CLL) patients. Results show that relational similarity can improve prediction over multiple baselines, for example a 5% incremental over long-short term memory baseline in terms of area under precision-recall curve.
Rare Disease Detection by Sequence Modeling with Generative Adversarial Networks
Jul 01, 2019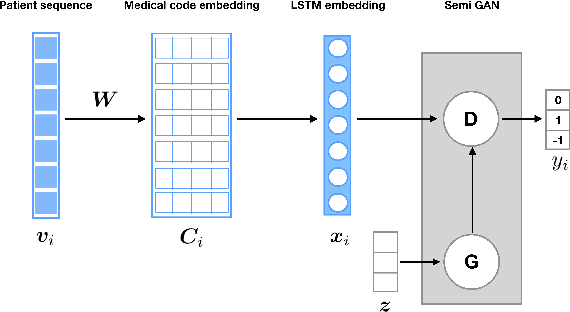
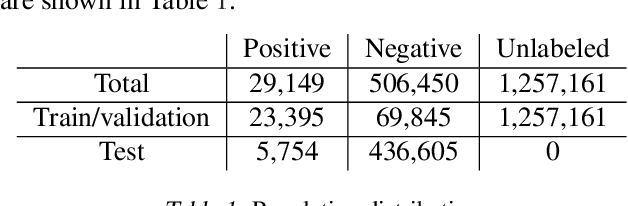
Abstract:Rare diseases affecting 350 million individuals are commonly associated with delay in diagnosis or misdiagnosis. To improve those patients' outcome, rare disease detection is an important task for identifying patients with rare conditions based on longitudinal medical claims. In this paper, we present a deep learning method for detecting patients with exocrine pancreatic insufficiency (EPI) (a rare disease). The contribution includes 1) a large longitudinal study using 7 years medical claims from 1.8 million patients including 29,149 EPI patients, 2) a new deep learning model using generative adversarial networks (GANs) to boost rare disease class, and also leveraging recurrent neural networks to model patient sequence data, 3) an accurate prediction with 0.56 PR-AUC which outperformed benchmark models in terms of precision and recall.
Modeling Treatment Delays for Patients using Feature Label Pairs in a Time Series
Dec 03, 2018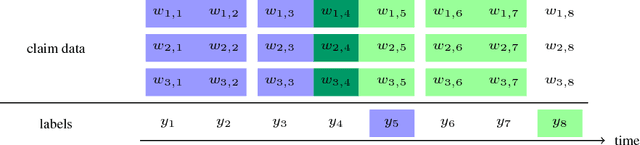
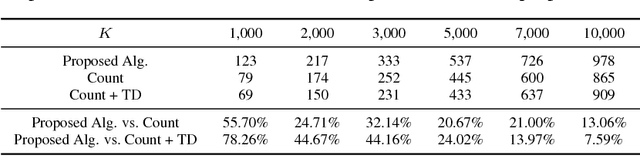
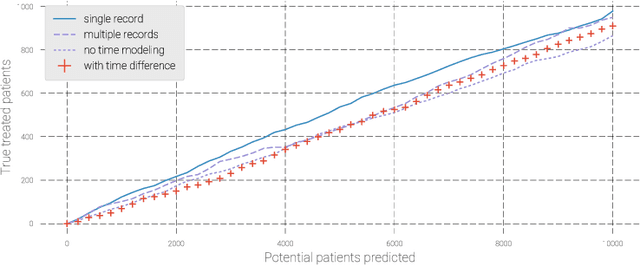
Abstract:Pharmaceutical targeting is one of key inputs for making sales and marketing strategy planning. Targeting list is built on predicting physician's sales potential of certain type of patient. In this paper, we present a time-sensitive targeting framework leveraging time series model to predict patient's disease and treatment progression. We create time features by extracting service history within a certain period, and record whether the event happens in a look-forward period. Such feature-label pairs are examined across all time periods and all patients to train a model. It keeps the inherent order of services and evaluates features associated to the imminent future, which contribute to improved accuracy.
Semi-supervised Rare Disease Detection Using Generative Adversarial Network
Dec 03, 2018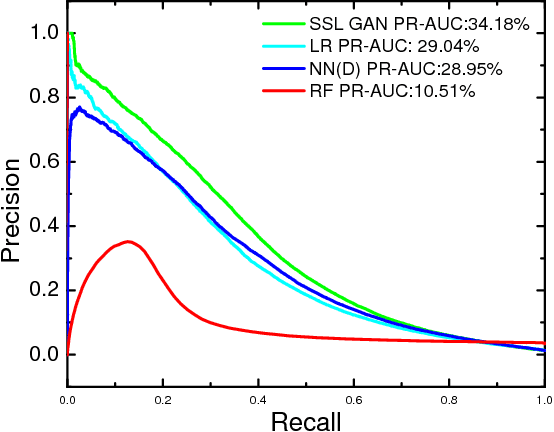
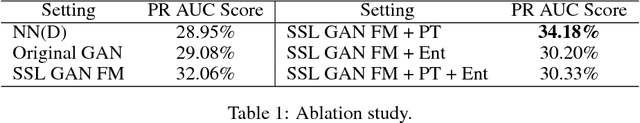
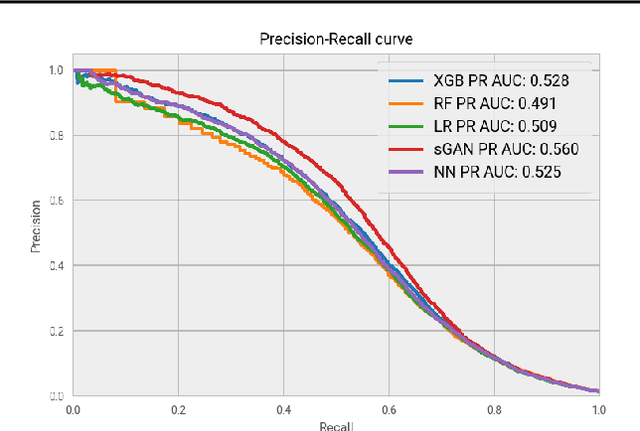
Abstract:Rare diseases affect a relatively small number of people, which limits investment in research for treatments and cures. Developing an efficient method for rare disease detection is a crucial first step towards subsequent clinical research. In this paper, we present a semi-supervised learning framework for rare disease detection using generative adversarial networks. Our method takes advantage of the large amount of unlabeled data for disease detection and achieves the best results in terms of precision-recall score compared to baseline techniques.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge