Dennis Shung
Learning over von Mises-Fisher Distributions via a Wasserstein-like Geometry
Apr 19, 2025Abstract:We introduce a novel, geometry-aware distance metric for the family of von Mises-Fisher (vMF) distributions, which are fundamental models for directional data on the unit hypersphere. Although the vMF distribution is widely employed in a variety of probabilistic learning tasks involving spherical data, principled tools for comparing vMF distributions remain limited, primarily due to the intractability of normalization constants and the absence of suitable geometric metrics. Motivated by the theory of optimal transport, we propose a Wasserstein-like distance that decomposes the discrepancy between two vMF distributions into two interpretable components: a geodesic term capturing the angular separation between mean directions, and a variance-like term quantifying differences in concentration parameters. The derivation leverages a Gaussian approximation in the high-concentration regime to yield a tractable, closed-form expression that respects the intrinsic spherical geometry. We show that the proposed distance exhibits desirable theoretical properties and induces a latent geometric structure on the space of non-degenerate vMF distributions. As a primary application, we develop the efficient algorithms for vMF mixture reduction, enabling structure-preserving compression of mixture models in high-dimensional settings. Empirical results on synthetic datasets and real-world high-dimensional embeddings, including biomedical sentence representations and deep visual features, demonstrate the effectiveness of the proposed geometry in distinguishing distributions and supporting interpretable inference. This work expands the statistical toolbox for directional data analysis by introducing a tractable, transport-inspired distance tailored to the geometry of the hypersphere.
Integrating Expert Judgment and Algorithmic Decision Making: An Indistinguishability Framework
Oct 11, 2024


Abstract:We introduce a novel framework for human-AI collaboration in prediction and decision tasks. Our approach leverages human judgment to distinguish inputs which are algorithmically indistinguishable, or "look the same" to any feasible predictive algorithm. We argue that this framing clarifies the problem of human-AI collaboration in prediction and decision tasks, as experts often form judgments by drawing on information which is not encoded in an algorithm's training data. Algorithmic indistinguishability yields a natural test for assessing whether experts incorporate this kind of "side information", and further provides a simple but principled method for selectively incorporating human feedback into algorithmic predictions. We show that this method provably improves the performance of any feasible algorithmic predictor and precisely quantify this improvement. We demonstrate the utility of our framework in a case study of emergency room triage decisions, where we find that although algorithmic risk scores are highly competitive with physicians, there is strong evidence that physician judgments provide signal which could not be replicated by any predictive algorithm. This insight yields a range of natural decision rules which leverage the complementary strengths of human experts and predictive algorithms.
Assessing the Usability of GutGPT: A Simulation Study of an AI Clinical Decision Support System for Gastrointestinal Bleeding Risk
Dec 06, 2023Abstract:Applications of large language models (LLMs) like ChatGPT have potential to enhance clinical decision support through conversational interfaces. However, challenges of human-algorithmic interaction and clinician trust are poorly understood. GutGPT, a LLM for gastrointestinal (GI) bleeding risk prediction and management guidance, was deployed in clinical simulation scenarios alongside the electronic health record (EHR) with emergency medicine physicians, internal medicine physicians, and medical students to evaluate its effect on physician acceptance and trust in AI clinical decision support systems (AI-CDSS). GutGPT provides risk predictions from a validated machine learning model and evidence-based answers by querying extracted clinical guidelines. Participants were randomized to GutGPT and an interactive dashboard, or the interactive dashboard and a search engine. Surveys and educational assessments taken before and after measured technology acceptance and content mastery. Preliminary results showed mixed effects on acceptance after using GutGPT compared to the dashboard or search engine but appeared to improve content mastery based on simulation performance. Overall, this study demonstrates LLMs like GutGPT could enhance effective AI-CDSS if implemented optimally and paired with interactive interfaces.
Auditing for Human Expertise
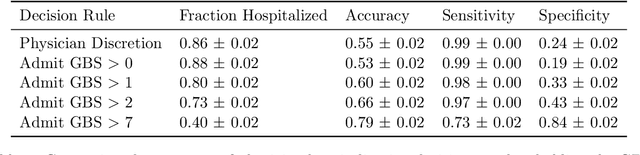
Jun 02, 2023Abstract:High-stakes prediction tasks (e.g., patient diagnosis) are often handled by trained human experts. A common source of concern about automation in these settings is that experts may exercise intuition that is difficult to model and/or have access to information (e.g., conversations with a patient) that is simply unavailable to a would-be algorithm. This raises a natural question whether human experts add value which could not be captured by an algorithmic predictor. We develop a statistical framework under which we can pose this question as a natural hypothesis test. Indeed, as our framework highlights, detecting human expertise is more subtle than simply comparing the accuracy of expert predictions to those made by a particular learning algorithm. Instead, we propose a simple procedure which tests whether expert predictions are statistically independent from the outcomes of interest after conditioning on the available inputs (`features'). A rejection of our test thus suggests that human experts may add value to any algorithm trained on the available data, and has direct implications for whether human-AI `complementarity' is achievable in a given prediction task. We highlight the utility of our procedure using admissions data collected from the emergency department of a large academic hospital system, where we show that physicians' admit/discharge decisions for patients with acute gastrointestinal bleeding (AGIB) appear to be incorporating information not captured in a standard algorithmic screening tool. This is despite the fact that the screening tool is arguably more accurate than physicians' discretionary decisions, highlighting that -- even absent normative concerns about accountability or interpretability -- accuracy is insufficient to justify algorithmic automation.
MURAL: An Unsupervised Random Forest-Based Embedding for Electronic Health Record Data
Nov 19, 2021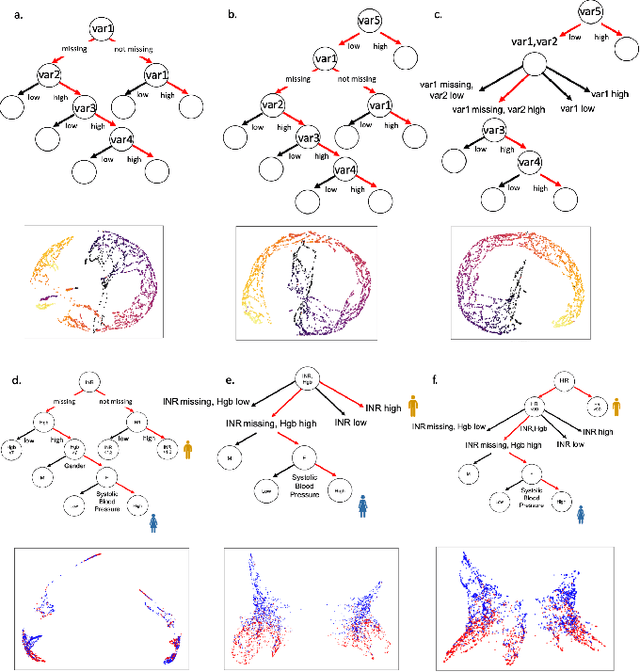
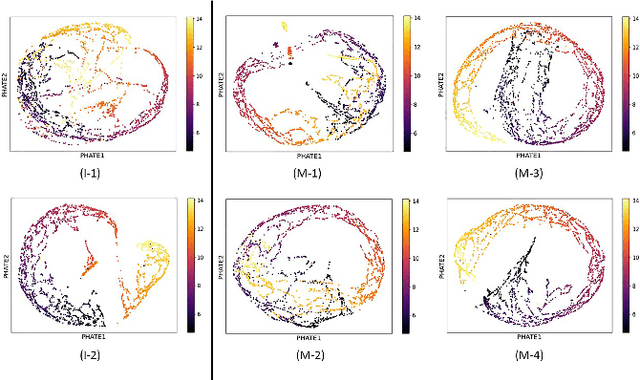
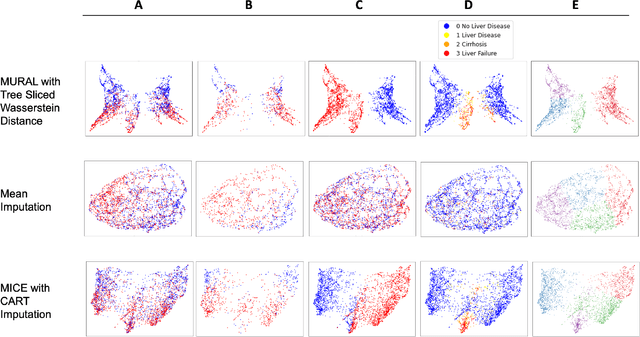

Abstract:A major challenge in embedding or visualizing clinical patient data is the heterogeneity of variable types including continuous lab values, categorical diagnostic codes, as well as missing or incomplete data. In particular, in EHR data, some variables are {\em missing not at random (MNAR)} but deliberately not collected and thus are a source of information. For example, lab tests may be deemed necessary for some patients on the basis of suspected diagnosis, but not for others. Here we present the MURAL forest -- an unsupervised random forest for representing data with disparate variable types (e.g., categorical, continuous, MNAR). MURAL forests consist of a set of decision trees where node-splitting variables are chosen at random, such that the marginal entropy of all other variables is minimized by the split. This allows us to also split on MNAR variables and discrete variables in a way that is consistent with the continuous variables. The end goal is to learn the MURAL embedding of patients using average tree distances between those patients. These distances can be fed to nonlinear dimensionality reduction method like PHATE to derive visualizable embeddings. While such methods are ubiquitous in continuous-valued datasets (like single cell RNA-sequencing) they have not been used extensively in mixed variable data. We showcase the use of our method on one artificial and two clinical datasets. We show that using our approach, we can visualize and classify data more accurately than competing approaches. Finally, we show that MURAL can also be used to compare cohorts of patients via the recently proposed tree-sliced Wasserstein distances.
Embedding Signals on Knowledge Graphs with Unbalanced Diffusion Earth Mover's Distance
Jul 26, 2021



Abstract:In modern relational machine learning it is common to encounter large graphs that arise via interactions or similarities between observations in many domains. Further, in many cases the target entities for analysis are actually signals on such graphs. We propose to compare and organize such datasets of graph signals by using an earth mover's distance (EMD) with a geodesic cost over the underlying graph. Typically, EMD is computed by optimizing over the cost of transporting one probability distribution to another over an underlying metric space. However, this is inefficient when computing the EMD between many signals. Here, we propose an unbalanced graph earth mover's distance that efficiently embeds the unbalanced EMD on an underlying graph into an $L^1$ space, whose metric we call unbalanced diffusion earth mover's distance (UDEMD). This leads us to an efficient nearest neighbors kernel over many signals defined on a large graph. Next, we show how this gives distances between graph signals that are robust to noise. Finally, we apply this to organizing patients based on clinical notes who are modelled as signals on the SNOMED-CT medical knowledge graph, embedding lymphoblast cells modeled as signals on a gene graph, and organizing genes modeled as signals over a large peripheral blood mononuclear (PBMC) cell graph. In each case, we show that UDEMD-based embeddings find accurate distances that are highly efficient compared to other methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge