Daniel Hernández-Lobato
Robust multi-task boosting using clustering and local ensembling
Feb 15, 2026Abstract:Multi-Task Learning (MTL) aims to boost predictive performance by sharing information across related tasks, yet conventional methods often suffer from negative transfer when unrelated or noisy tasks are forced to share representations. We propose Robust Multi-Task Boosting using Clustering and Local Ensembling (RMB-CLE), a principled MTL framework that integrates error-based task clustering with local ensembling. Unlike prior work that assumes fixed clusters or hand-crafted similarity metrics, RMB-CLE derives inter-task similarity directly from cross-task errors, which admit a risk decomposition into functional mismatch and irreducible noise, providing a theoretically grounded mechanism to prevent negative transfer. Tasks are grouped adaptively via agglomerative clustering, and within each cluster, a local ensemble enables robust knowledge sharing while preserving task-specific patterns. Experiments show that RMB-CLE recovers ground-truth clusters in synthetic data and consistently outperforms multi-task, single-task, and pooling-based ensemble methods across diverse real-world and synthetic benchmarks. These results demonstrate that RMB-CLE is not merely a combination of clustering and boosting but a general and scalable framework that establishes a new basis for robust multi-task learning.
Improving the Linearized Laplace Approximation via Quadratic Approximations
Feb 03, 2026Abstract:Deep neural networks (DNNs) often produce overconfident out-of-distribution predictions, motivating Bayesian uncertainty quantification. The Linearized Laplace Approximation (LLA) achieves this by linearizing the DNN and applying Laplace inference to the resulting model. Importantly, the linear model is also used for prediction. We argue this linearization in the posterior may degrade fidelity to the true Laplace approximation. To alleviate this problem, without increasing significantly the computational cost, we propose the Quadratic Laplace Approximation (QLA). QLA approximates each second order factor in the approximate Laplace log-posterior using a rank-one factor obtained via efficient power iterations. QLA is expected to yield a posterior precision closer to that of the full Laplace without forming the full Hessian, which is typically intractable. For prediction, QLA also uses the linearized model. Empirically, QLA yields modest yet consistent uncertainty estimation improvements over LLA on five regression datasets.
Scalable Linearized Laplace Approximation via Surrogate Neural Kernel
Jan 29, 2026Abstract:We introduce a scalable method to approximate the kernel of the Linearized Laplace Approximation (LLA). For this, we use a surrogate deep neural network (DNN) that learns a compact feature representation whose inner product replicates the Neural Tangent Kernel (NTK). This avoids the need to compute large Jacobians. Training relies solely on efficient Jacobian-vector products, allowing to compute predictive uncertainty on large-scale pre-trained DNNs. Experimental results show similar or improved uncertainty estimation and calibration compared to existing LLA approximations. Notwithstanding, biasing the learned kernel significantly enhances out-of-distribution detection. This remarks the benefits of the proposed method for finding better kernels than the NTK in the context of LLA to compute prediction uncertainty given a pre-trained DNN.
Robust-Multi-Task Gradient Boosting
Jul 15, 2025Abstract:Multi-task learning (MTL) has shown effectiveness in exploiting shared information across tasks to improve generalization. MTL assumes tasks share similarities that can improve performance. In addition, boosting algorithms have demonstrated exceptional performance across diverse learning problems, primarily due to their ability to focus on hard-to-learn instances and iteratively reduce residual errors. This makes them a promising approach for learning multi-task problems. However, real-world MTL scenarios often involve tasks that are not well-aligned (known as outlier or adversarial tasks), which do not share beneficial similarities with others and can, in fact, deteriorate the performance of the overall model. To overcome this challenge, we propose Robust-Multi-Task Gradient Boosting (R-MTGB), a novel boosting framework that explicitly models and adapts to task heterogeneity during training. R-MTGB structures the learning process into three sequential blocks: (1) learning shared patterns, (2) partitioning tasks into outliers and non-outliers with regularized parameters, and (3) fine-tuning task-specific predictors. This architecture enables R-MTGB to automatically detect and penalize outlier tasks while promoting effective knowledge transfer among related tasks. Our method integrates these mechanisms seamlessly within gradient boosting, allowing robust handling of noisy or adversarial tasks without sacrificing accuracy. Extensive experiments on both synthetic benchmarks and real-world datasets demonstrate that our approach successfully isolates outliers, transfers knowledge, and consistently reduces prediction errors for each task individually, and achieves overall performance gains across all tasks. These results highlight robustness, adaptability, and reliable convergence of R-MTGB in challenging MTL environments.
Fixed-Mean Gaussian Processes for Post-hoc Bayesian Deep Learning
Dec 05, 2024Abstract:Recently, there has been an increasing interest in performing post-hoc uncertainty estimation about the predictions of pre-trained deep neural networks (DNNs). Given a pre-trained DNN via back-propagation, these methods enhance the original network by adding output confidence measures, such as error bars, without compromising its initial accuracy. In this context, we introduce a novel family of sparse variational Gaussian processes (GPs), where the posterior mean is fixed to any continuous function when using a universal kernel. Specifically, we fix the mean of this GP to the output of the pre-trained DNN, allowing our approach to effectively fit the GP's predictive variances to estimate the DNN prediction uncertainty. Our approach leverages variational inference (VI) for efficient stochastic optimization, with training costs that remain independent of the number of training points, scaling efficiently to large datasets such as ImageNet. The proposed method, called fixed mean GP (FMGP), is architecture-agnostic, relying solely on the pre-trained model's outputs to adjust the predictive variances. Experimental results demonstrate that FMGP improves both uncertainty estimation and computational efficiency when compared to state-of-the-art methods.
Alpha Entropy Search for New Information-based Bayesian Optimization
Nov 25, 2024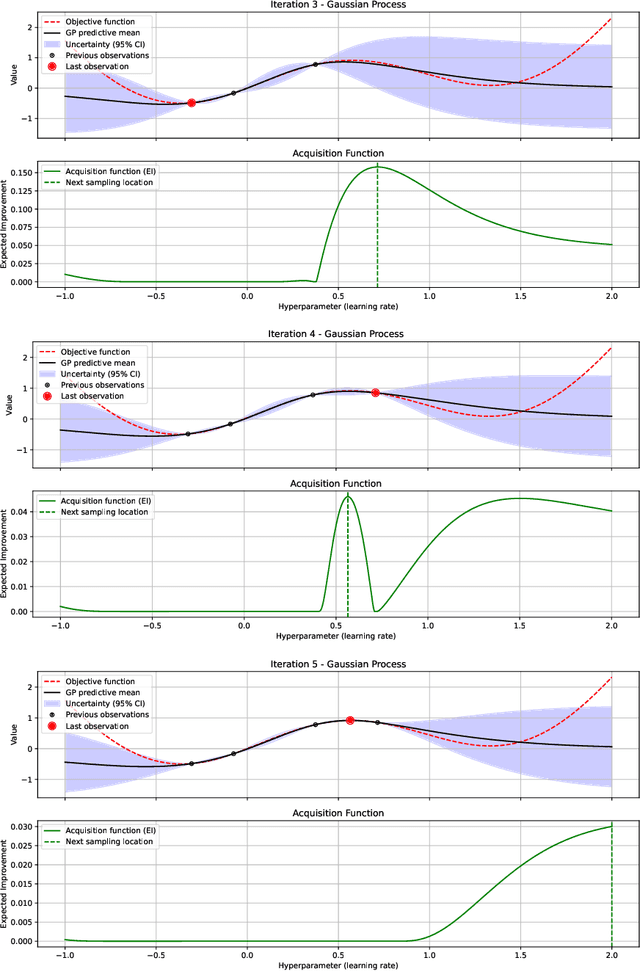
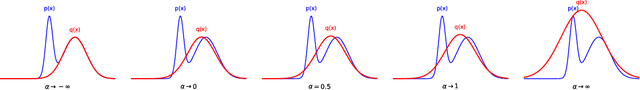
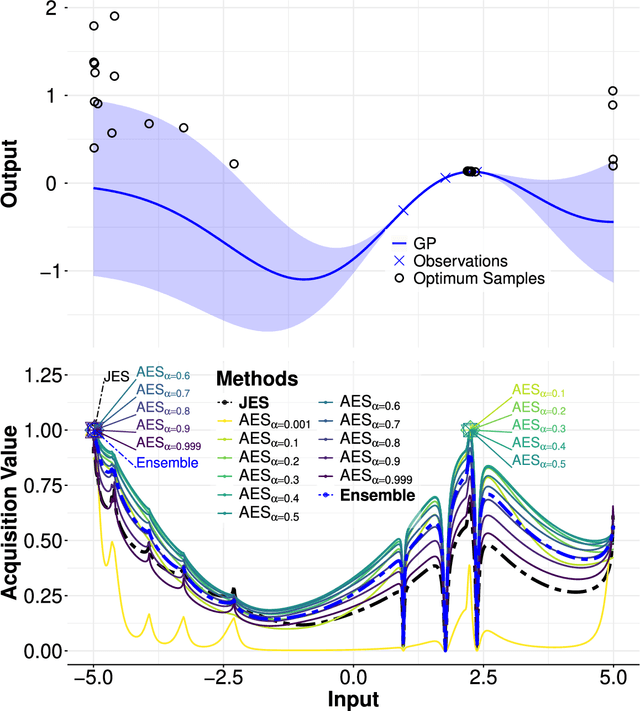
Abstract:Bayesian optimization (BO) methods based on information theory have obtained state-of-the-art results in several tasks. These techniques heavily rely on the Kullback-Leibler (KL) divergence to compute the acquisition function. In this work, we introduce a novel information-based class of acquisition functions for BO called Alpha Entropy Search (AES). AES is based on the {\alpha}-divergence, that generalizes the KL divergence. Iteratively, AES selects the next evaluation point as the one whose associated target value has the highest level of the dependency with respect to the location and associated value of the global maximum of the optimization problem. Dependency is measured in terms of the {\alpha}-divergence, as an alternative to the KL divergence. Intuitively, this favors the evaluation of the objective function at the most informative points about the global maximum. The {\alpha}-divergence has a free parameter {\alpha}, which determines the behavior of the divergence, trading-off evaluating differences between distributions at a single mode, and evaluating differences globally. Therefore, different values of {\alpha} result in different acquisition functions. AES acquisition lacks a closed-form expression. However, we propose an efficient and accurate approximation using a truncated Gaussian distribution. In practice, the value of {\alpha} can be chosen by the practitioner, but here we suggest to use a combination of acquisition functions obtained by simultaneously considering a range of values of {\alpha}. We provide an implementation of AES in BOTorch and we evaluate its performance in both synthetic, benchmark and real-world experiments involving the tuning of the hyper-parameters of a deep neural network. These experiments show that the performance of AES is competitive with respect to other information-based acquisition functions such as JES, MES or PES.
Deep Transformed Gaussian Processes
Nov 02, 2023Abstract:Transformed Gaussian Processes (TGPs) are stochastic processes specified by transforming samples from the joint distribution from a prior process (typically a GP) using an invertible transformation; increasing the flexibility of the base process. Furthermore, they achieve competitive results compared with Deep Gaussian Processes (DGPs), which are another generalization constructed by a hierarchical concatenation of GPs. In this work, we propose a generalization of TGPs named Deep Transformed Gaussian Processes (DTGPs), which follows the trend of concatenating layers of stochastic processes. More precisely, we obtain a multi-layer model in which each layer is a TGP. This generalization implies an increment of flexibility with respect to both TGPs and DGPs. Exact inference in such a model is intractable. However, we show that one can use variational inference to approximate the required computations yielding a straightforward extension of the popular DSVI inference algorithm Salimbeni et al (2017). The experiments conducted evaluate the proposed novel DTGPs in multiple regression datasets, achieving good scalability and performance.
Variational Linearized Laplace Approximation for Bayesian Deep Learning
Feb 24, 2023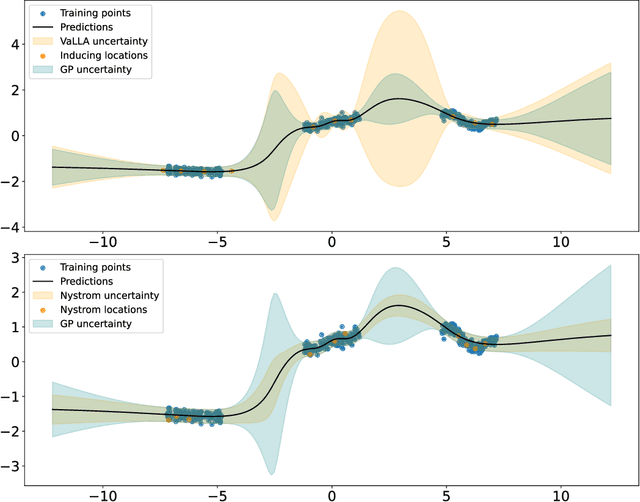
Abstract:Pre-trained deep neural networks can be adapted to perform uncertainty estimation by transforming them into Bayesian neural networks via methods such as Laplace approximation (LA) or its linearized form (LLA), among others. To make these methods more tractable, the generalized Gauss-Newton (GGN) approximation is often used. However, due to complex inefficiency difficulties, both LA and LLA rely on further approximations, such as Kronecker-factored or diagonal approximate GGN matrices, which can affect the results. To address these issues, we propose a new method for scaling LLA using a variational sparse Gaussian Process (GP) approximation based on the dual RKHS of GPs. Our method retains the predictive mean of the original model while allowing for efficient stochastic optimization and scalability in both the number of parameters and the size of the training dataset. Moreover, its training cost is independent of the number of training points, improving over previously existing methods. Our preliminary experiments indicate that it outperforms already existing efficient variants of LLA, such as accelerated LLA (ELLA), based on the Nystr\"om approximation.
Correcting Model Bias with Sparse Implicit Processes
Aug 08, 2022
Abstract:Model selection in machine learning (ML) is a crucial part of the Bayesian learning procedure. Model choice may impose strong biases on the resulting predictions, which can hinder the performance of methods such as Bayesian neural networks and neural samplers. On the other hand, newly proposed approaches for Bayesian ML exploit features of approximate inference in function space with implicit stochastic processes (a generalization of Gaussian processes). The approach of Sparse Implicit Processes (SIP) is particularly successful in this regard, since it is fully trainable and achieves flexible predictions. Here, we expand on the original experiments to show that SIP is capable of correcting model bias when the data generating mechanism differs strongly from the one implied by the model. We use synthetic datasets to show that SIP is capable of providing predictive distributions that reflect the data better than the exact predictions of the initial, but wrongly assumed model.
Deep Variational Implicit Processes
Jun 14, 2022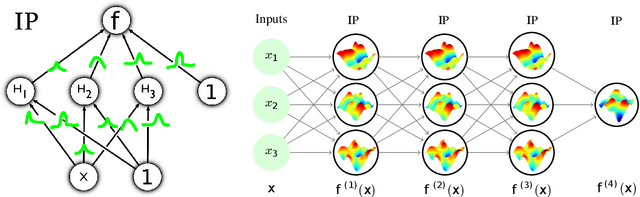
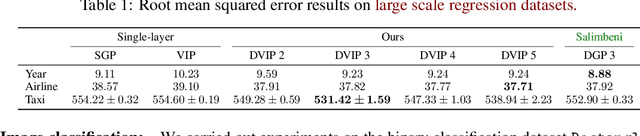
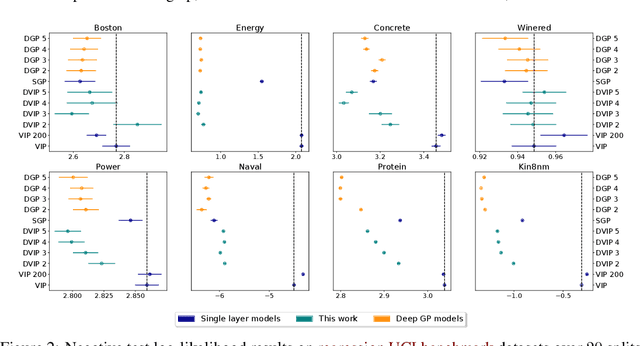

Abstract:Implicit processes (IPs) are a generalization of Gaussian processes (GPs). IPs may lack a closed-form expression but are easy to sample from. Examples include, among others, Bayesian neural networks or neural samplers. IPs can be used as priors over functions, resulting in flexible models with well-calibrated prediction uncertainty estimates. Methods based on IPs usually carry out function-space approximate inference, which overcomes some of the difficulties of parameter-space approximate inference. Nevertheless, the approximations employed often limit the expressiveness of the final model, resulting, \emph{e.g.}, in a Gaussian predictive distribution, which can be restrictive. We propose here a multi-layer generalization of IPs called the Deep Variational Implicit process (DVIP). This generalization is similar to that of deep GPs over GPs, but it is more flexible due to the use of IPs as the prior distribution over the latent functions. We describe a scalable variational inference algorithm for training DVIP and show that it outperforms previous IP-based methods and also deep GPs. We support these claims via extensive regression and classification experiments. We also evaluate DVIP on large datasets with up to several million data instances to illustrate its good scalability and performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge