Christopher Weight
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
AI Age Discrepancy: A Novel Parameter for Frailty Assessment in Kidney Tumor Patients
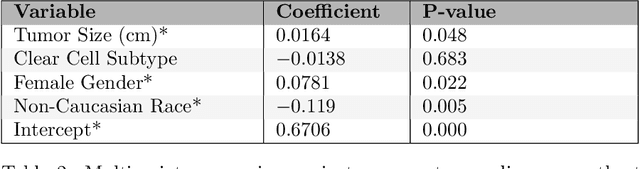
Jul 02, 2024Abstract:Kidney cancer is a global health concern, and accurate assessment of patient frailty is crucial for optimizing surgical outcomes. This paper introduces AI Age Discrepancy, a novel metric derived from machine learning analysis of preoperative abdominal CT scans, as a potential indicator of frailty and postoperative risk in kidney cancer patients. This retrospective study of 599 patients from the 2023 Kidney Tumor Segmentation (KiTS) challenge dataset found that a higher AI Age Discrepancy is significantly associated with longer hospital stays and lower overall survival rates, independent of established factors. This suggests that AI Age Discrepancy may provide valuable insights into patient frailty and could thus inform clinical decision-making in kidney cancer treatment.
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023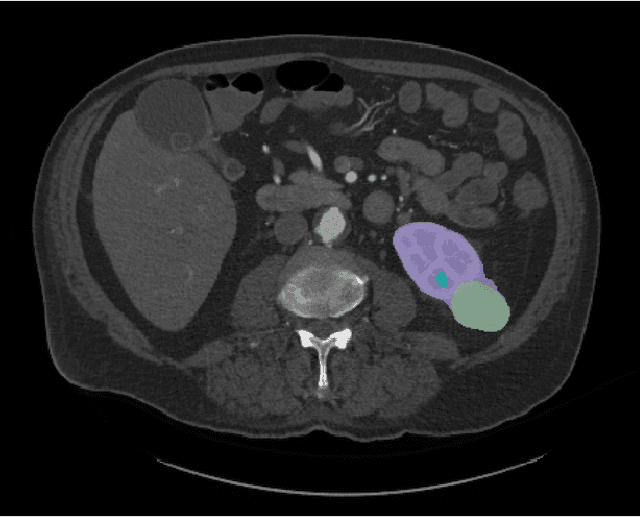
Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 Challenge
Dec 02, 2019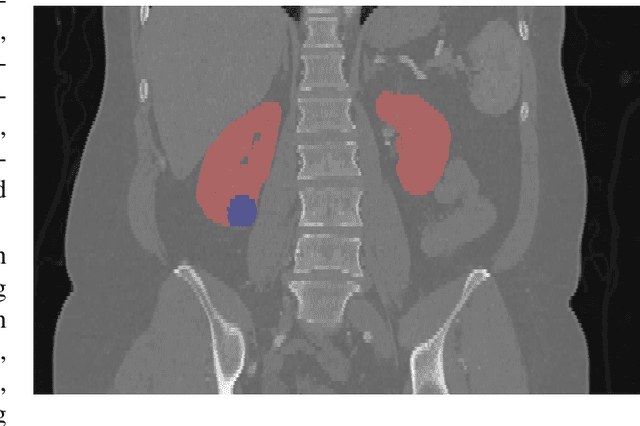
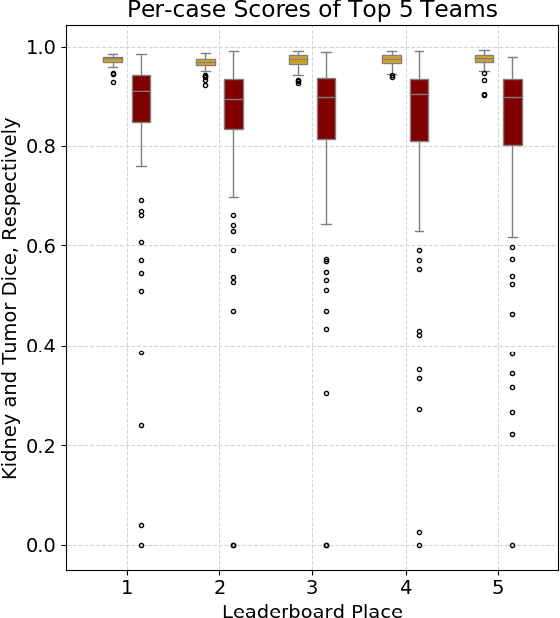
Abstract:There is a large body of literature linking anatomic and geometric characteristics of kidney tumors to perioperative and oncologic outcomes. Semantic segmentation of these tumors and their host kidneys is a promising tool for quantitatively characterizing these lesions, but its adoption is limited due to the manual effort required to produce high-quality 3D segmentations of these structures. Recently, methods based on deep learning have shown excellent results in automatic 3D segmentation, but they require large datasets for training, and there remains little consensus on which methods perform best. The 2019 Kidney and Kidney Tumor Segmentation challenge (KiTS19) was a competition held in conjunction with the 2019 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) which sought to address these issues and stimulate progress on this automatic segmentation problem. A training set of 210 cross sectional CT images with kidney tumors was publicly released with corresponding semantic segmentation masks. 106 teams from five continents used this data to develop automated systems to predict the true segmentation masks on a test set of 90 CT images for which the corresponding ground truth segmentations were kept private. These predictions were scored and ranked according to their average So rensen-Dice coefficient between the kidney and tumor across all 90 cases. The winning team achieved a Dice of 0.974 for kidney and 0.851 for tumor, approaching the inter-annotator performance on kidney (0.983) but falling short on tumor (0.923). This challenge has now entered an "open leaderboard" phase where it serves as a challenging benchmark in 3D semantic segmentation.
The Role of Publicly Available Data in MICCAI Papers from 2014 to 2018
Aug 12, 2019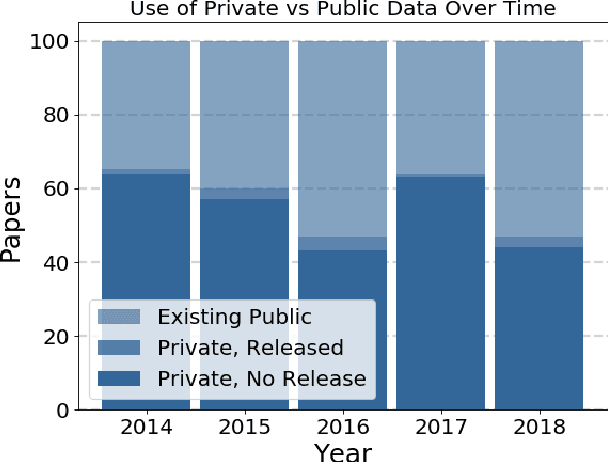
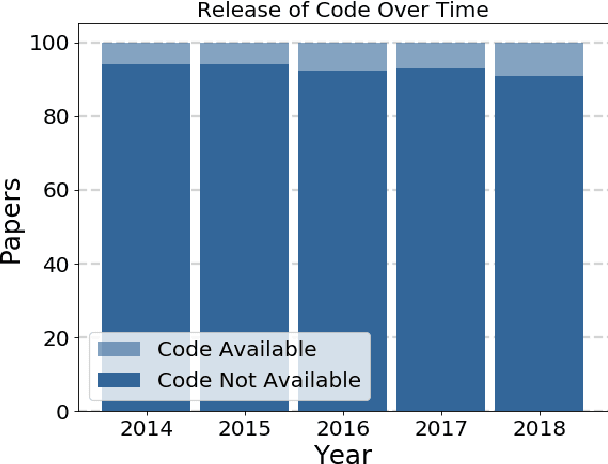
Abstract:Widely-used public benchmarks are of huge importance to computer vision and machine learning research, especially with the computational resources required to reproduce state of the art results quickly becoming untenable. In medical image computing, the wide variety of image modalities and problem formulations yields a huge task-space for benchmarks to cover, and thus the widespread adoption of standard benchmarks has been slow, and barriers to releasing medical data exacerbate this issue. In this paper, we examine the role that publicly available data has played in MICCAI papers from the past five years. We find that more than half of these papers are based on private data alone, although this proportion seems to be decreasing over time. Additionally, we observed that after controlling for open access publication and the release of code, papers based on public data were cited over 60% more per year than their private-data counterparts. Further, we found that more than 20% of papers using public data did not provide a citation to the dataset or associated manuscript, highlighting the "second-rate" status that data contributions often take compared to theoretical ones. We conclude by making recommendations for MICCAI policies which could help to better incentivise data sharing and move the field toward more efficient and reproducible science.
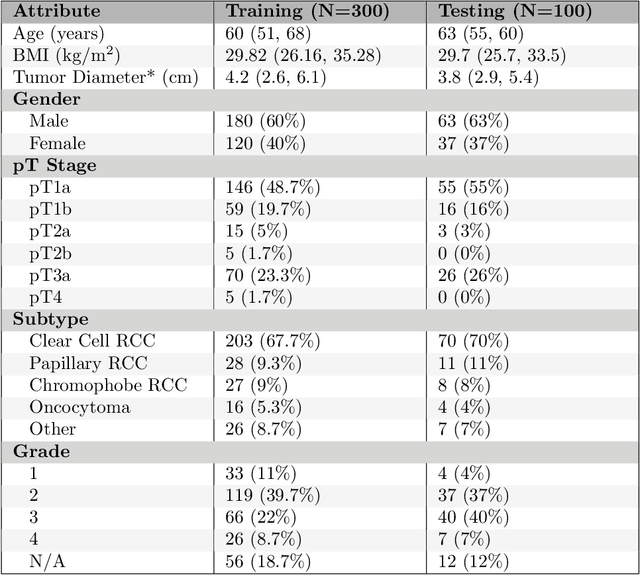
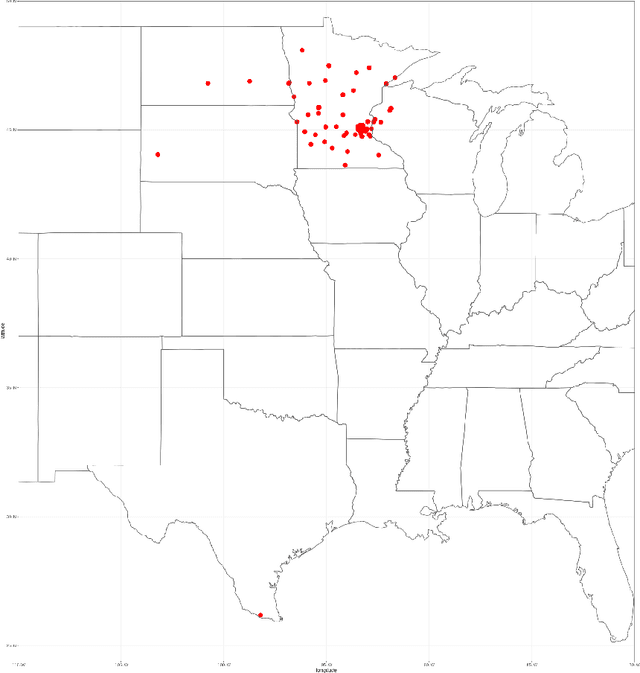
The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes
Mar 31, 2019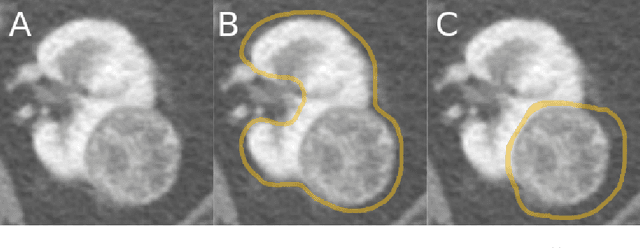
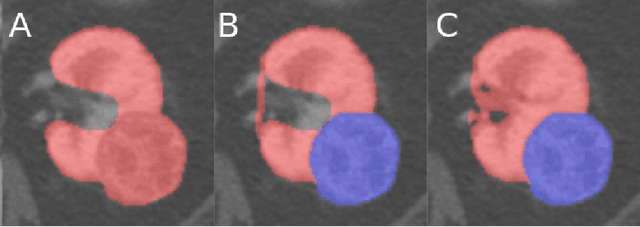
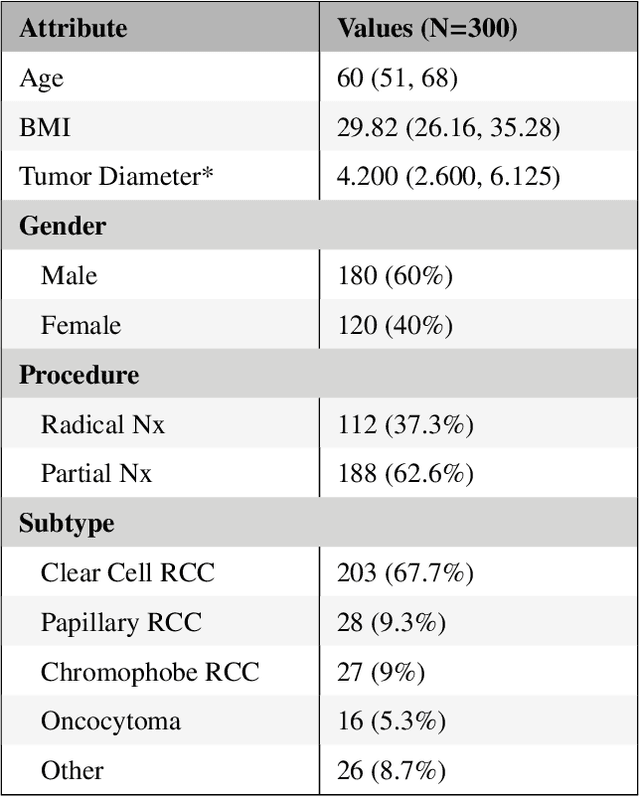
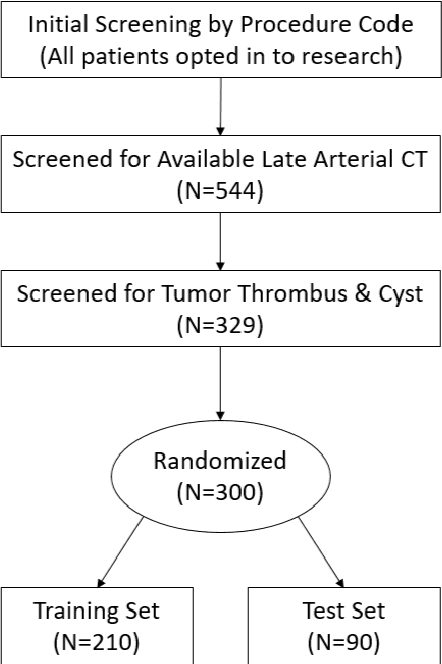
Abstract:The morphometry of a kidney tumor revealed by contrast-enhanced Computed Tomography (CT) imaging is an important factor in clinical decision making surrounding the lesion's diagnosis and treatment. Quantitative study of the relationship between kidney tumor morphology and clinical outcomes is difficult due to data scarcity and the laborious nature of manually quantifying imaging predictors. Automatic semantic segmentation of kidneys and kidney tumors is a promising tool towards automatically quantifying a wide array of morphometric features, but no sizeable annotated dataset is currently available to train models for this task. We present the KiTS19 challenge dataset: A collection of multi-phase CT imaging, segmentation masks, and comprehensive clinical outcomes for 300 patients who underwent nephrectomy for kidney tumors at our center between 2010 and 2018. 210 (70%) of these patients were selected at random as the training set for the 2019 MICCAI KiTS Kidney Tumor Segmentation Challenge and have been released publicly. With the presence of clinical context and surgical outcomes, this data can serve not only for benchmarking semantic segmentation models, but also for developing and studying biomarkers which make use of the imaging and semantic segmentation masks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge