Chengzhang Zhu
A multitask framework for automated interpretation of multi-frame right upper quadrant ultrasound in clinical decision support
Jan 17, 2026Abstract:Ultrasound is a cornerstone of emergency and hepatobiliary imaging, yet its interpretation remains highly operator-dependent and time-sensitive. Here, we present a multitask vision-language agent (VLM) developed to assist with comprehensive right upper quadrant (RUQ) ultrasound interpretation across the full diagnostic workflow. The system was trained on a large, multi-center dataset comprising a primary cohort from Johns Hopkins Medical Institutions (9,189 cases, 594,099 images) and externally validated on cohorts from Stanford University (108 cases, 3,240 images) and a major Chinese medical center (257 cases, 3,178 images). Built on the Qwen2.5-VL-7B architecture, the agent integrates frame-level visual understanding with report-grounded language reasoning to perform three tasks: (i) classification of 18 hepatobiliary and gallbladder conditions, (ii) generation of clinically coherent diagnostic reports, and (iii) surgical decision support based on ultrasound findings and clinical data. The model achieved high diagnostic accuracy across all tasks, generated reports that were indistinguishable from expert-written versions in blinded evaluations, and demonstrated superior factual accuracy and information density on content-based metrics. The agent further identified patients requiring cholecystectomy with high precision, supporting real-time decision-making. These results highlight the potential of generalist vision-language models to improve diagnostic consistency, reporting efficiency, and surgical triage in real-world ultrasound practice.
The Evolution and Future Perspectives of Artificial Intelligence Generated Content
Dec 02, 2024



Abstract:Artificial intelligence generated content (AIGC), a rapidly advancing technology, is transforming content creation across domains, such as text, images, audio, and video. Its growing potential has attracted more and more researchers and investors to explore and expand its possibilities. This review traces AIGC's evolution through four developmental milestones-ranging from early rule-based systems to modern transfer learning models-within a unified framework that highlights how each milestone contributes uniquely to content generation. In particular, the paper employs a common example across all milestones to illustrate the capabilities and limitations of methods within each phase, providing a consistent evaluation of AIGC methodologies and their development. Furthermore, this paper addresses critical challenges associated with AIGC and proposes actionable strategies to mitigate them. This study aims to guide researchers and practitioners in selecting and optimizing AIGC models to enhance the quality and efficiency of content creation across diverse domains.
Leveraging CORAL-Correlation Consistency Network for Semi-Supervised Left Atrium MRI Segmentation
Oct 21, 2024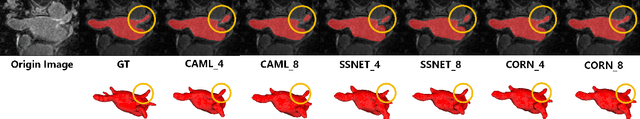
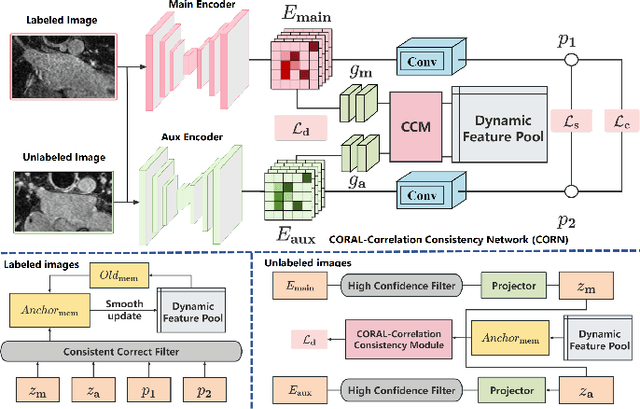
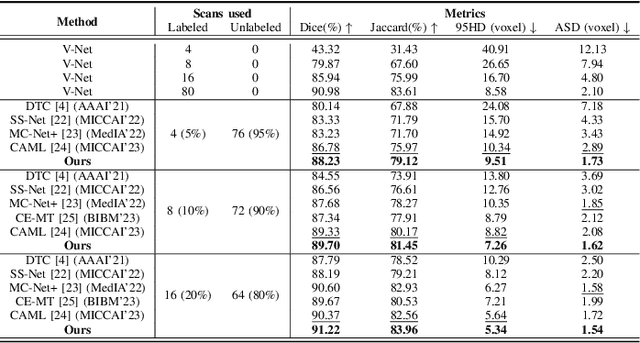
Abstract:Semi-supervised learning (SSL) has been widely used to learn from both a few labeled images and many unlabeled images to overcome the scarcity of labeled samples in medical image segmentation. Most current SSL-based segmentation methods use pixel values directly to identify similar features in labeled and unlabeled data. They usually fail to accurately capture the intricate attachment structures in the left atrium, such as the areas of inconsistent density or exhibit outward curvatures, adding to the complexity of the task. In this paper, we delve into this issue and introduce an effective solution, CORAL(Correlation-Aligned)-Correlation Consistency Network (CORN), to capture the global structure shape and local details of Left Atrium. Diverging from previous methods focused on each local pixel value, the CORAL-Correlation Consistency Module (CCM) in the CORN leverages second-order statistical information to capture global structural features by minimizing the distribution discrepancy between labeled and unlabeled samples in feature space. Yet, direct construction of features from unlabeled data frequently results in ``Sample Selection Bias'', leading to flawed supervision. We thus further propose the Dynamic Feature Pool (DFP) for the CCM, which utilizes a confidence-based filtering strategy to remove incorrectly selected features and regularize both teacher and student models by constraining the similarity matrix to be consistent. Extensive experiments on the Left Atrium dataset have shown that the proposed CORN outperforms previous state-of-the-art semi-supervised learning methods.
Active Learning in Brain Tumor Segmentation with Uncertainty Sampling, Annotation Redundancy Restriction, and Data Initialization
Feb 05, 2023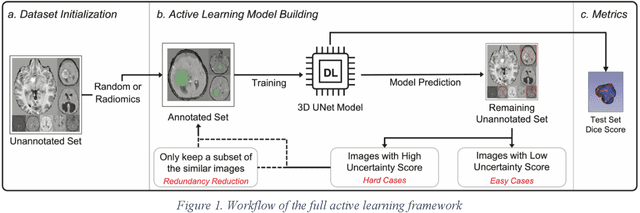
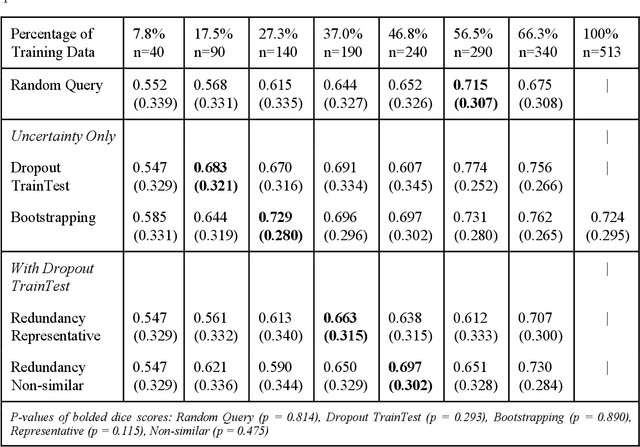
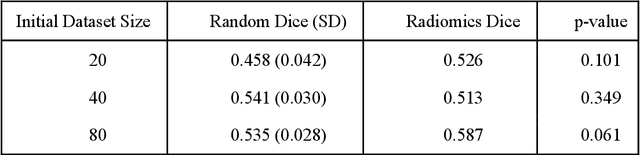
Abstract:Deep learning models have demonstrated great potential in medical 3D imaging, but their development is limited by the expensive, large volume of annotated data required. Active learning (AL) addresses this by training a model on a subset of the most informative data samples without compromising performance. We compared different AL strategies and propose a framework that minimizes the amount of data needed for state-of-the-art performance. 638 multi-institutional brain tumor MRI images were used to train a 3D U-net model and compare AL strategies. We investigated uncertainty sampling, annotation redundancy restriction, and initial dataset selection techniques. Uncertainty estimation techniques including Bayesian estimation with dropout, bootstrapping, and margins sampling were compared to random query. Strategies to avoid annotation redundancy by removing similar images within the to-be-annotated subset were considered as well. We determined the minimum amount of data necessary to achieve similar performance to the model trained on the full dataset ({\alpha} = 0.1). A variance-based selection strategy using radiomics to identify the initial training dataset is also proposed. Bayesian approximation with dropout at training and testing showed similar results to that of the full data model with less than 20% of the training data (p=0.293) compared to random query achieving similar performance at 56.5% of the training data (p=0.814). Annotation redundancy restriction techniques achieved state-of-the-art performance at approximately 40%-50% of the training data. Radiomics dataset initialization had higher Dice with initial dataset sizes of 20 and 80 images, but improvements were not significant. In conclusion, we investigated various AL strategies with dropout uncertainty estimation achieving state-of-the-art performance with the least annotated data.
Table2Vec: Automated Universal Representation Learning to Encode All-round Data DNA for Benchmarkable and Explainable Enterprise Data Science
Dec 03, 2021



Abstract:Enterprise data typically involves multiple heterogeneous data sources and external data that respectively record business activities, transactions, customer demographics, status, behaviors, interactions and communications with the enterprise, and the consumption and feedback of its products, services, production, marketing, operations, and management, etc. A critical challenge in enterprise data science is to enable an effective whole-of-enterprise data understanding and data-driven discovery and decision-making on all-round enterprise DNA. We introduce a neural encoder Table2Vec for automated universal representation learning of entities such as customers from all-round enterprise DNA with automated data characteristics analysis and data quality augmentation. The learned universal representations serve as representative and benchmarkable enterprise data genomes and can be used for enterprise-wide and domain-specific learning tasks. Table2Vec integrates automated universal representation learning on low-quality enterprise data and downstream learning tasks. We illustrate Table2Vec in characterizing all-round customer data DNA in an enterprise on complex heterogeneous multi-relational big tables to build universal customer vector representations. The learned universal representation of each customer is all-round, representative and benchmarkable to support both enterprise-wide and domain-specific learning goals and tasks in enterprise data science. Table2Vec significantly outperforms the existing shallow, boosting and deep learning methods typically used for enterprise analytics. We further discuss the research opportunities, directions and applications of automated universal enterprise representation and learning and the learned enterprise data DNA for automated, all-purpose, whole-of-enterprise and ethical machine learning and data science.
Personalized next-best action recommendation with multi-party interaction learning for automated decision-making
Aug 19, 2021



Abstract:Automated next-best action recommendation for each customer in a sequential, dynamic and interactive context has been widely needed in natural, social and business decision-making. Personalized next-best action recommendation must involve past, current and future customer demographics and circumstances (states) and behaviors, long-range sequential interactions between customers and decision-makers, multi-sequence interactions between states, behaviors and actions, and their reactions to their counterpart's actions. No existing modeling theories and tools, including Markovian decision processes, user and behavior modeling, deep sequential modeling, and personalized sequential recommendation, can quantify such complex decision-making on a personal level. We take a data-driven approach to learn the next-best actions for personalized decision-making by a reinforced coupled recurrent neural network (CRN). CRN represents multiple coupled dynamic sequences of a customer's historical and current states, responses to decision-makers' actions, decision rewards to actions, and learns long-term multi-sequence interactions between parties (customer and decision-maker). Next-best actions are then recommended on each customer at a time point to change their state for an optimal decision-making objective. Our study demonstrates the potential of personalized deep learning of multi-sequence interactions and automated dynamic intervention for personalized decision-making in complex systems.
Sensing Anomalies like Humans: A Hominine Framework to Detect Abnormal Events from Unlabeled Videos
Aug 04, 2021
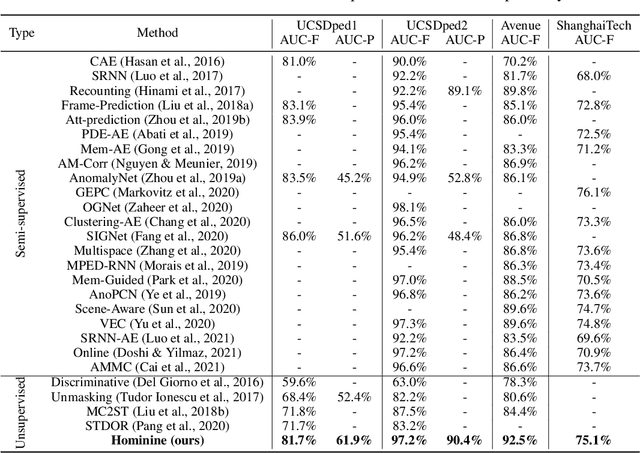
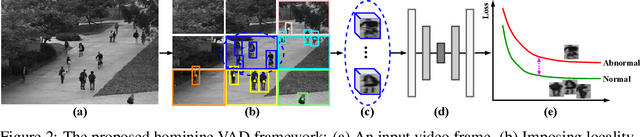

Abstract:Video anomaly detection (VAD) has constantly been a vital topic in video analysis. As anomalies are often rare, it is typically addressed under a semi-supervised setup, which requires a training set with pure normal videos. To avoid exhausted manual labeling, we are inspired by how humans sense anomalies and propose a hominine framework that enables both unsupervised and end-to-end VAD. The framework is based on two key observations: 1) Human perception is usually local, i.e. focusing on local foreground and its context when sensing anomalies. Thus, we propose to impose locality-awareness by localizing foreground with generic knowledge, and a region localization strategy is designed to exploit local context. 2) Frequently-occurred events will mould humans' definition of normality, which motivates us to devise a surrogate training paradigm. It trains a deep neural network (DNN) to learn a surrogate task with unlabeled videos, and frequently-occurred events will play a dominant role in "moulding" the DNN. In this way, a training loss gap will automatically manifest rarely-seen novel events as anomalies. For implementation, we explore various surrogate tasks as well as both classic and emerging DNN models. Extensive evaluations on commonly-used VAD benchmarks justify the framework's applicability to different surrogate tasks or DNN models, and demonstrate its astonishing effectiveness: It not only outperforms existing unsupervised solutions by a wide margin (8% to 10% AUROC gain), but also achieves comparable or even superior performance to state-of-the-art semi-supervised counterparts.
Unsupervised Heterogeneous Coupling Learning for Categorical Representation
Jul 21, 2020
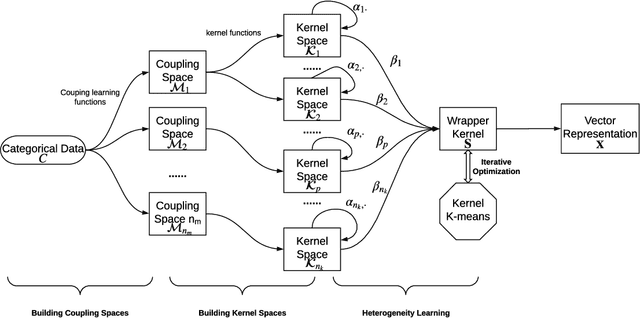

Abstract:Complex categorical data is often hierarchically coupled with heterogeneous relationships between attributes and attribute values and the couplings between objects. Such value-to-object couplings are heterogeneous with complementary and inconsistent interactions and distributions. Limited research exists on unlabeled categorical data representations, ignores the heterogeneous and hierarchical couplings, underestimates data characteristics and complexities, and overuses redundant information, etc. The deep representation learning of unlabeled categorical data is challenging, overseeing such value-to-object couplings, complementarity and inconsistency, and requiring large data, disentanglement, and high computational power. This work introduces a shallow but powerful UNsupervised heTerogeneous couplIng lEarning (UNTIE) approach for representing coupled categorical data by untying the interactions between couplings and revealing heterogeneous distributions embedded in each type of couplings. UNTIE is efficiently optimized w.r.t. a kernel k-means objective function for unsupervised representation learning of heterogeneous and hierarchical value-to-object couplings. Theoretical analysis shows that UNTIE can represent categorical data with maximal separability while effectively represent heterogeneous couplings and disclose their roles in categorical data. The UNTIE-learned representations make significant performance improvement against the state-of-the-art categorical representations and deep representation models on 25 categorical data sets with diversified characteristics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge