Chetan Bettegowda
Active Learning in Brain Tumor Segmentation with Uncertainty Sampling, Annotation Redundancy Restriction, and Data Initialization
Feb 05, 2023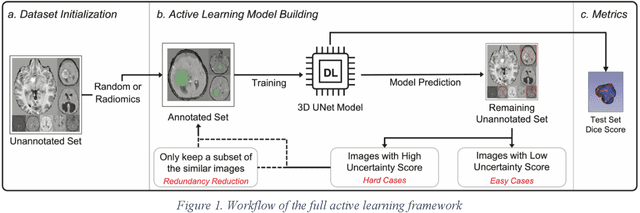
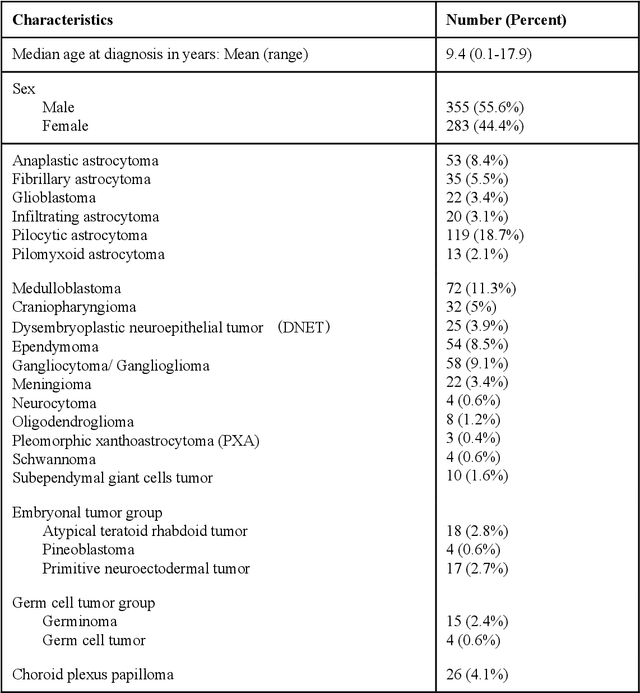
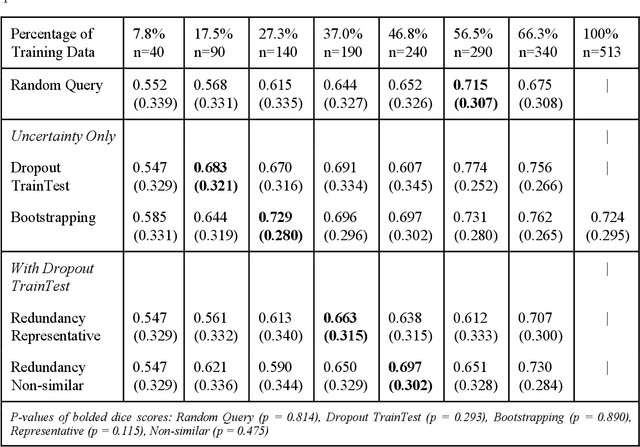
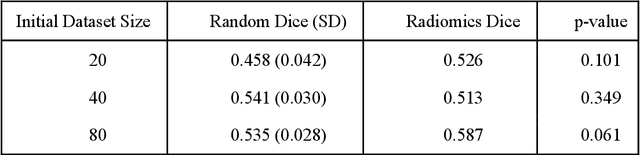
Abstract:Deep learning models have demonstrated great potential in medical 3D imaging, but their development is limited by the expensive, large volume of annotated data required. Active learning (AL) addresses this by training a model on a subset of the most informative data samples without compromising performance. We compared different AL strategies and propose a framework that minimizes the amount of data needed for state-of-the-art performance. 638 multi-institutional brain tumor MRI images were used to train a 3D U-net model and compare AL strategies. We investigated uncertainty sampling, annotation redundancy restriction, and initial dataset selection techniques. Uncertainty estimation techniques including Bayesian estimation with dropout, bootstrapping, and margins sampling were compared to random query. Strategies to avoid annotation redundancy by removing similar images within the to-be-annotated subset were considered as well. We determined the minimum amount of data necessary to achieve similar performance to the model trained on the full dataset ({\alpha} = 0.1). A variance-based selection strategy using radiomics to identify the initial training dataset is also proposed. Bayesian approximation with dropout at training and testing showed similar results to that of the full data model with less than 20% of the training data (p=0.293) compared to random query achieving similar performance at 56.5% of the training data (p=0.814). Annotation redundancy restriction techniques achieved state-of-the-art performance at approximately 40%-50% of the training data. Radiomics dataset initialization had higher Dice with initial dataset sizes of 20 and 80 images, but improvements were not significant. In conclusion, we investigated various AL strategies with dropout uncertainty estimation achieving state-of-the-art performance with the least annotated data.
Multiparametric Deep Learning and Radiomics for Tumor Grading and Treatment Response Assessment of Brain Cancer: Preliminary Results
Jun 10, 2019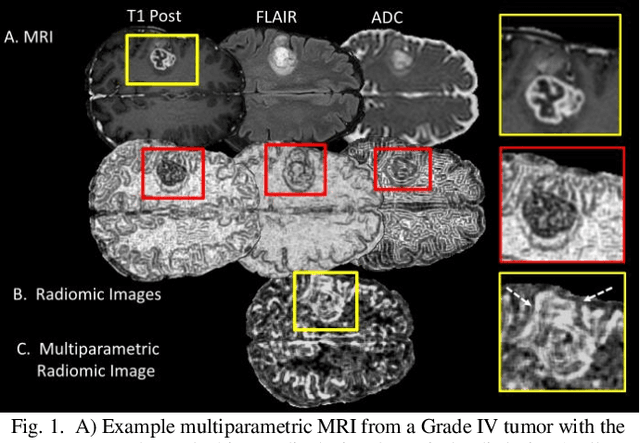
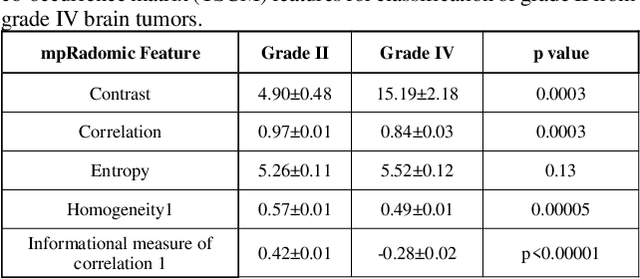
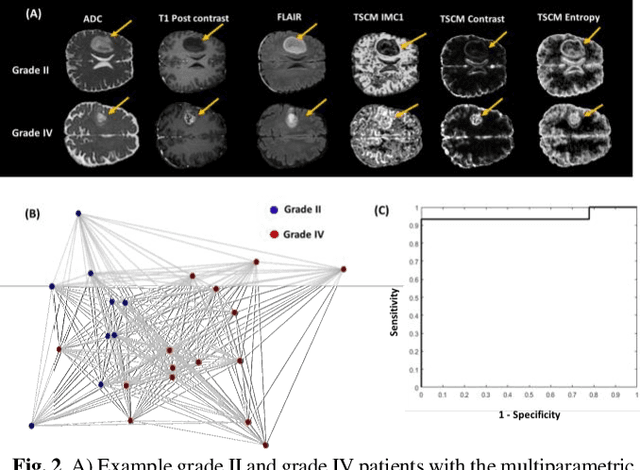
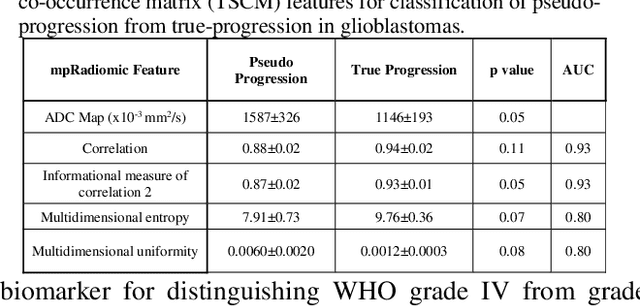
Abstract:Radiomics is an exciting new area of texture research for extracting quantitative and morphological characteristics of pathological tissue. However, to date, only single images have been used for texture analysis. We have extended radiomic texture methods to use multiparametric (mp) data to get more complete information from all the images. These mpRadiomic methods could potentially provide a platform for stratification of tumor grade as well as assessment of treatment response in brain tumors. In brain, multiparametric MRI (mpMRI) are based on contrast enhanced T1-weighted imaging (T1WI), T2WI, Fluid Attenuated Inversion Recovery (FLAIR), Diffusion Weighted Imaging (DWI) and Perfusion Weighted Imaging (PWI). Therefore, we applied our multiparametric radiomic framework (mpRadiomic) on 24 patients with brain tumors (8 grade II and 16 grade IV). The mpRadiomic framework classified grade IV tumors from grade II tumors with a sensitivity and specificity of 93% and 100%, respectively, with an AUC of 0.95. For treatment response, the mpRadiomic framework classified pseudo-progression from true-progression with an AUC of 0.93. In conclusion, the mpRadiomic analysis was able to effectively capture the multiparametric brain MRI texture and could be used as potential biomarkers for distinguishing grade IV from grade II tumors as well as determining true-progression from pseudo-progression.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge