Vishwa S. Parekh
Towards Fair Medical AI: Adversarial Debiasing of 3D CT Foundation Embeddings
Feb 05, 2025



Abstract:Self-supervised learning has revolutionized medical imaging by enabling efficient and generalizable feature extraction from large-scale unlabeled datasets. Recently, self-supervised foundation models have been extended to three-dimensional (3D) computed tomography (CT) data, generating compact, information-rich embeddings with 1408 features that achieve state-of-the-art performance on downstream tasks such as intracranial hemorrhage detection and lung cancer risk forecasting. However, these embeddings have been shown to encode demographic information, such as age, sex, and race, which poses a significant risk to the fairness of clinical applications. In this work, we propose a Variation Autoencoder (VAE) based adversarial debiasing framework to transform these embeddings into a new latent space where demographic information is no longer encoded, while maintaining the performance of critical downstream tasks. We validated our approach on the NLST lung cancer screening dataset, demonstrating that the debiased embeddings effectively eliminate multiple encoded demographic information and improve fairness without compromising predictive accuracy for lung cancer risk at 1-year and 2-year intervals. Additionally, our approach ensures the embeddings are robust against adversarial bias attacks. These results highlight the potential of adversarial debiasing techniques to ensure fairness and equity in clinical applications of self-supervised 3D CT embeddings, paving the way for their broader adoption in unbiased medical decision-making.
Demographic Predictability in 3D CT Foundation Embeddings
Nov 28, 2024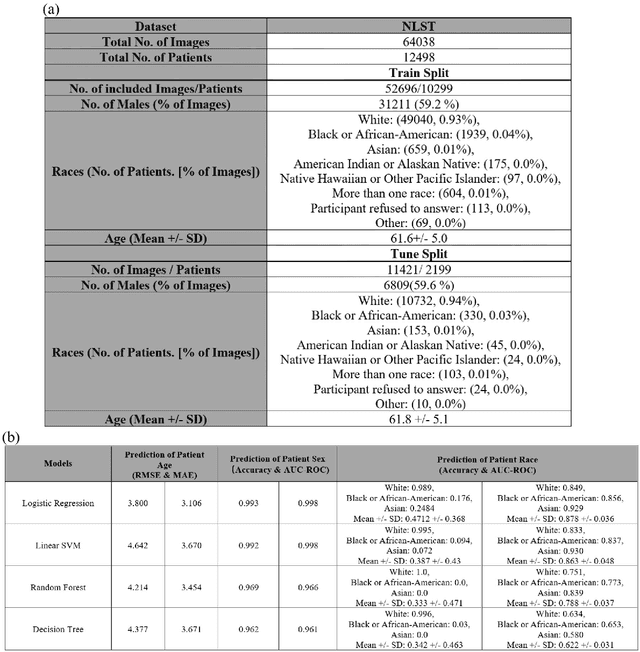

Abstract:Self-supervised foundation models have recently been successfully extended to encode three-dimensional (3D) computed tomography (CT) images, with excellent performance across several downstream tasks, such as intracranial hemorrhage detection and lung cancer risk forecasting. However, as self-supervised models learn from complex data distributions, questions arise concerning whether these embeddings capture demographic information, such as age, sex, or race. Using the National Lung Screening Trial (NLST) dataset, which contains 3D CT images and demographic data, we evaluated a range of classifiers: softmax regression, linear regression, linear support vector machine, random forest, and decision tree, to predict sex, race, and age of the patients in the images. Our results indicate that the embeddings effectively encoded age and sex information, with a linear regression model achieving a root mean square error (RMSE) of 3.8 years for age prediction and a softmax regression model attaining an AUC of 0.998 for sex classification. Race prediction was less effective, with an AUC of 0.878. These findings suggest a detailed exploration into the information encoded in self-supervised learning frameworks is needed to help ensure fair, responsible, and patient privacy-protected healthcare AI.
Expanding the Horizon: Enabling Hybrid Quantum Transfer Learning for Long-Tailed Chest X-Ray Classification
Apr 30, 2024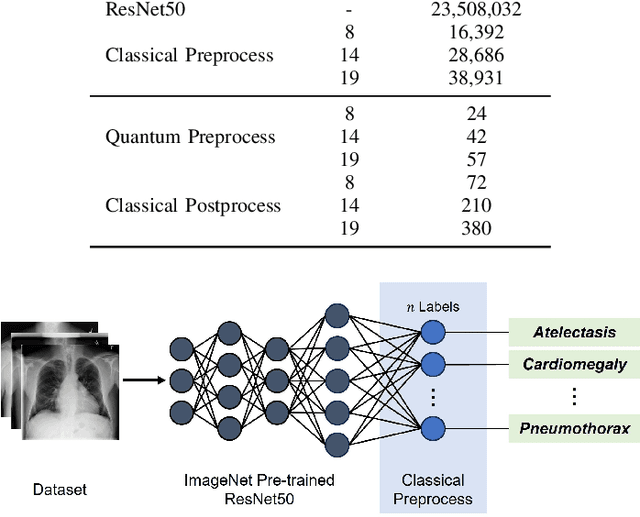
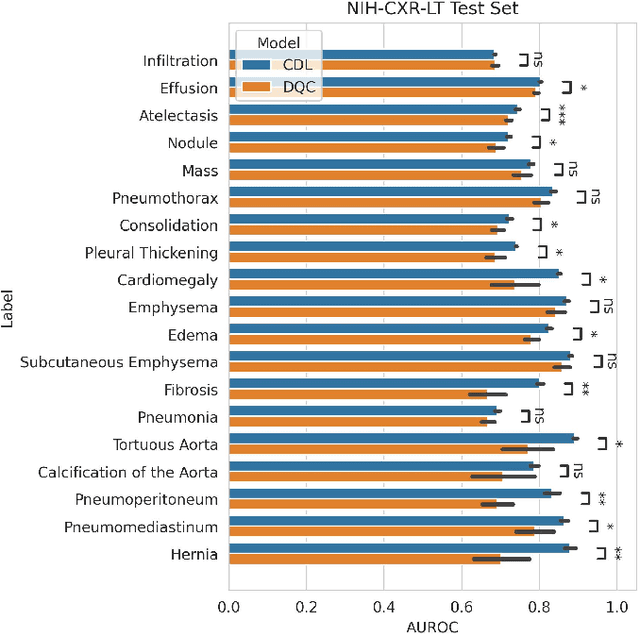
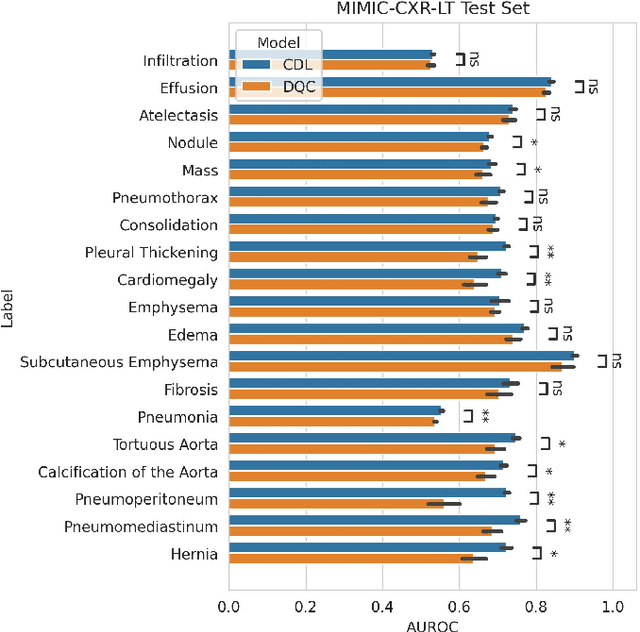
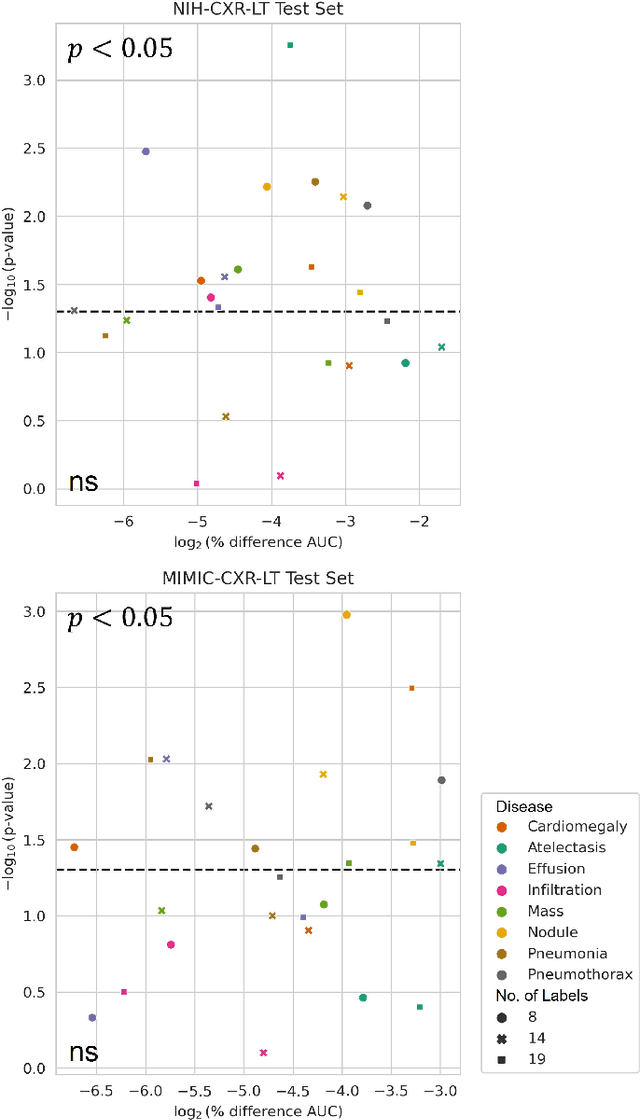
Abstract:Quantum machine learning (QML) has the potential for improving the multi-label classification of rare, albeit critical, diseases in large-scale chest x-ray (CXR) datasets due to theoretical quantum advantages over classical machine learning (CML) in sample efficiency and generalizability. While prior literature has explored QML with CXRs, it has focused on binary classification tasks with small datasets due to limited access to quantum hardware and computationally expensive simulations. To that end, we implemented a Jax-based framework that enables the simulation of medium-sized qubit architectures with significant improvements in wall-clock time over current software offerings. We evaluated the performance of our Jax-based framework in terms of efficiency and performance for hybrid quantum transfer learning for long-tailed classification across 8, 14, and 19 disease labels using large-scale CXR datasets. The Jax-based framework resulted in up to a 58% and 95% speed-up compared to PyTorch and TensorFlow implementations, respectively. However, compared to CML, QML demonstrated slower convergence and an average AUROC of 0.70, 0.73, and 0.74 for the classification of 8, 14, and 19 CXR disease labels. In comparison, the CML models had an average AUROC of 0.77, 0.78, and 0.80 respectively. In conclusion, our work presents an accessible implementation of hybrid quantum transfer learning for long-tailed CXR classification with a computationally efficient Jax-based framework.
Improving Multi-Center Generalizability of GAN-Based Fat Suppression using Federated Learning
Apr 10, 2024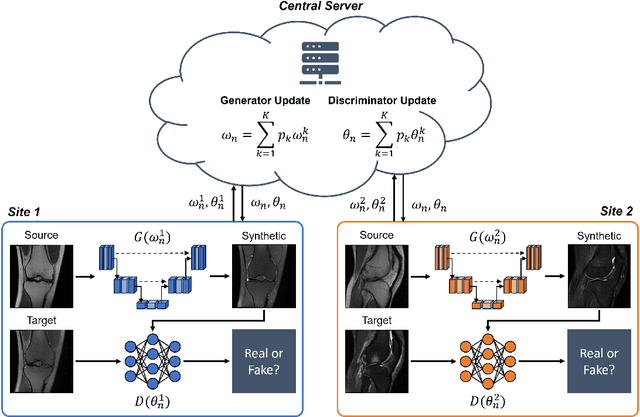
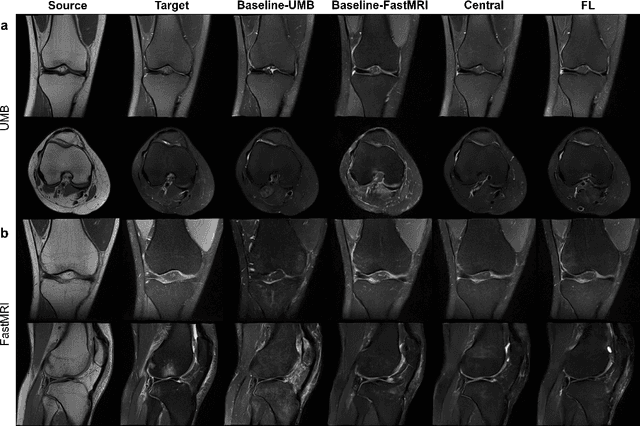
Abstract:Generative Adversarial Network (GAN)-based synthesis of fat suppressed (FS) MRIs from non-FS proton density sequences has the potential to accelerate acquisition of knee MRIs. However, GANs trained on single-site data have poor generalizability to external data. We show that federated learning can improve multi-center generalizability of GANs for synthesizing FS MRIs, while facilitating privacy-preserving multi-institutional collaborations.
Anytime, Anywhere, Anyone: Investigating the Feasibility of Segment Anything Model for Crowd-Sourcing Medical Image Annotations
Mar 22, 2024Abstract:Curating annotations for medical image segmentation is a labor-intensive and time-consuming task that requires domain expertise, resulting in "narrowly" focused deep learning (DL) models with limited translational utility. Recently, foundation models like the Segment Anything Model (SAM) have revolutionized semantic segmentation with exceptional zero-shot generalizability across various domains, including medical imaging, and hold a lot of promise for streamlining the annotation process. However, SAM has yet to be evaluated in a crowd-sourced setting to curate annotations for training 3D DL segmentation models. In this work, we explore the potential of SAM for crowd-sourcing "sparse" annotations from non-experts to generate "dense" segmentation masks for training 3D nnU-Net models, a state-of-the-art DL segmentation model. Our results indicate that while SAM-generated annotations exhibit high mean Dice scores compared to ground-truth annotations, nnU-Net models trained on SAM-generated annotations perform significantly worse than nnU-Net models trained on ground-truth annotations ($p<0.001$, all).
Hidden in Plain Sight: Undetectable Adversarial Bias Attacks on Vulnerable Patient Populations
Feb 08, 2024Abstract:The proliferation of artificial intelligence (AI) in radiology has shed light on the risk of deep learning (DL) models exacerbating clinical biases towards vulnerable patient populations. While prior literature has focused on quantifying biases exhibited by trained DL models, demographically targeted adversarial bias attacks on DL models and its implication in the clinical environment remains an underexplored field of research in medical imaging. In this work, we demonstrate that demographically targeted label poisoning attacks can introduce adversarial underdiagnosis bias in DL models and degrade performance on underrepresented groups without impacting overall model performance. Moreover, our results across multiple performance metrics and demographic groups like sex, age, and their intersectional subgroups indicate that a group's vulnerability to undetectable adversarial bias attacks is directly correlated with its representation in the model's training data.
One Copy Is All You Need: Resource-Efficient Streaming of Medical Imaging Data at Scale
Jul 01, 2023Abstract:Large-scale medical imaging datasets have accelerated development of artificial intelligence tools for clinical decision support. However, the large size of these datasets is a bottleneck for users with limited storage and bandwidth. Many users may not even require such large datasets as AI models are often trained on lower resolution images. If users could directly download at their desired resolution, storage and bandwidth requirements would significantly decrease. However, it is impossible to anticipate every users' requirements and impractical to store the data at multiple resolutions. What if we could store images at a single resolution but send them at different ones? We propose MIST, an open-source framework to operationalize progressive resolution for streaming medical images at multiple resolutions from a single high-resolution copy. We demonstrate that MIST can dramatically reduce imaging infrastructure inefficiencies for hosting and streaming medical images by >90%, while maintaining diagnostic quality for deep learning applications.
A framework for dynamically training and adapting deep reinforcement learning models to different, low-compute, and continuously changing radiology deployment environments
Jun 08, 2023



Abstract:While Deep Reinforcement Learning has been widely researched in medical imaging, the training and deployment of these models usually require powerful GPUs. Since imaging environments evolve rapidly and can be generated by edge devices, the algorithm is required to continually learn and adapt to changing environments, and adjust to low-compute devices. To this end, we developed three image coreset algorithms to compress and denoise medical images for selective experience replayed-based lifelong reinforcement learning. We implemented neighborhood averaging coreset, neighborhood sensitivity-based sampling coreset, and maximum entropy coreset on full-body DIXON water and DIXON fat MRI images. All three coresets produced 27x compression with excellent performance in localizing five anatomical landmarks: left knee, right trochanter, left kidney, spleen, and lung across both imaging environments. Maximum entropy coreset obtained the best performance of $11.97\pm 12.02$ average distance error, compared to the conventional lifelong learning framework's $19.24\pm 50.77$.
Multi-environment lifelong deep reinforcement learning for medical imaging
May 31, 2023Abstract:Deep reinforcement learning(DRL) is increasingly being explored in medical imaging. However, the environments for medical imaging tasks are constantly evolving in terms of imaging orientations, imaging sequences, and pathologies. To that end, we developed a Lifelong DRL framework, SERIL to continually learn new tasks in changing imaging environments without catastrophic forgetting. SERIL was developed using selective experience replay based lifelong learning technique for the localization of five anatomical landmarks in brain MRI on a sequence of twenty-four different imaging environments. The performance of SERIL, when compared to two baseline setups: MERT(multi-environment-best-case) and SERT(single-environment-worst-case) demonstrated excellent performance with an average distance of $9.90\pm7.35$ pixels from the desired landmark across all 120 tasks, compared to $10.29\pm9.07$ for MERT and $36.37\pm22.41$ for SERT($p<0.05$), demonstrating the excellent potential for continuously learning multiple tasks across dynamically changing imaging environments.
High-Throughput AI Inference for Medical Image Classification and Segmentation using Intelligent Streaming
May 24, 2023Abstract:As the adoption of AI systems within the clinical setup grows, limitations in bandwidth could create communication bottlenecks when streaming imaging data, leading to delays in patient diagnosis and treatment. As such, healthcare providers and AI vendors will require greater computational infrastructure, therefore dramatically increasing costs. To that end, we developed intelligent streaming, a state-of-the-art framework to enable accelerated, cost-effective, bandwidth-optimized, and computationally efficient AI inference for clinical decision making at scale. For classification, intelligent streaming reduced the data transmission by 99.01% and decoding time by 98.58%, while increasing throughput by 27.43x. For segmentation, our framework reduced data transmission by 90.32%, decoding time by 90.26%, while increasing throughput by 4.20x. Our work demonstrates that intelligent streaming results in faster turnaround times, and reduced overall cost of data and transmission, without negatively impacting clinical decision making using AI systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge