Guangyao Zheng
Towards Fair Medical AI: Adversarial Debiasing of 3D CT Foundation Embeddings
Feb 05, 2025



Abstract:Self-supervised learning has revolutionized medical imaging by enabling efficient and generalizable feature extraction from large-scale unlabeled datasets. Recently, self-supervised foundation models have been extended to three-dimensional (3D) computed tomography (CT) data, generating compact, information-rich embeddings with 1408 features that achieve state-of-the-art performance on downstream tasks such as intracranial hemorrhage detection and lung cancer risk forecasting. However, these embeddings have been shown to encode demographic information, such as age, sex, and race, which poses a significant risk to the fairness of clinical applications. In this work, we propose a Variation Autoencoder (VAE) based adversarial debiasing framework to transform these embeddings into a new latent space where demographic information is no longer encoded, while maintaining the performance of critical downstream tasks. We validated our approach on the NLST lung cancer screening dataset, demonstrating that the debiased embeddings effectively eliminate multiple encoded demographic information and improve fairness without compromising predictive accuracy for lung cancer risk at 1-year and 2-year intervals. Additionally, our approach ensures the embeddings are robust against adversarial bias attacks. These results highlight the potential of adversarial debiasing techniques to ensure fairness and equity in clinical applications of self-supervised 3D CT embeddings, paving the way for their broader adoption in unbiased medical decision-making.
Demographic Predictability in 3D CT Foundation Embeddings
Nov 28, 2024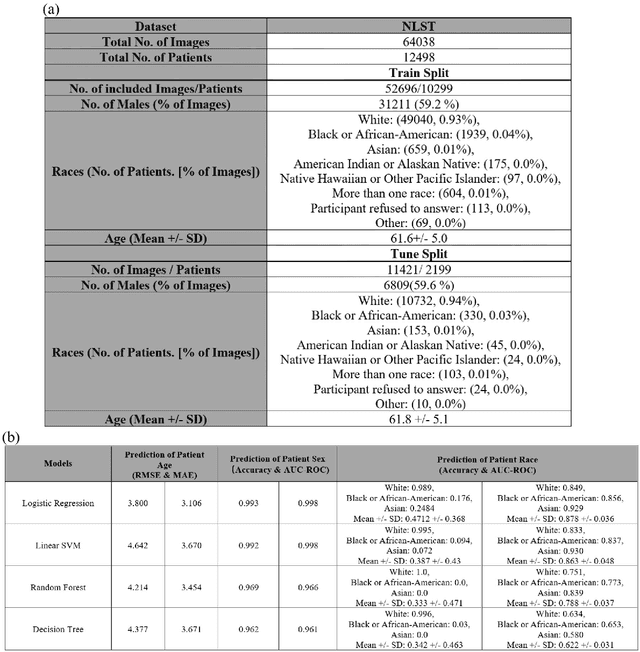

Abstract:Self-supervised foundation models have recently been successfully extended to encode three-dimensional (3D) computed tomography (CT) images, with excellent performance across several downstream tasks, such as intracranial hemorrhage detection and lung cancer risk forecasting. However, as self-supervised models learn from complex data distributions, questions arise concerning whether these embeddings capture demographic information, such as age, sex, or race. Using the National Lung Screening Trial (NLST) dataset, which contains 3D CT images and demographic data, we evaluated a range of classifiers: softmax regression, linear regression, linear support vector machine, random forest, and decision tree, to predict sex, race, and age of the patients in the images. Our results indicate that the embeddings effectively encoded age and sex information, with a linear regression model achieving a root mean square error (RMSE) of 3.8 years for age prediction and a softmax regression model attaining an AUC of 0.998 for sex classification. Race prediction was less effective, with an AUC of 0.878. These findings suggest a detailed exploration into the information encoded in self-supervised learning frameworks is needed to help ensure fair, responsible, and patient privacy-protected healthcare AI.
A framework for dynamically training and adapting deep reinforcement learning models to different, low-compute, and continuously changing radiology deployment environments
Jun 08, 2023



Abstract:While Deep Reinforcement Learning has been widely researched in medical imaging, the training and deployment of these models usually require powerful GPUs. Since imaging environments evolve rapidly and can be generated by edge devices, the algorithm is required to continually learn and adapt to changing environments, and adjust to low-compute devices. To this end, we developed three image coreset algorithms to compress and denoise medical images for selective experience replayed-based lifelong reinforcement learning. We implemented neighborhood averaging coreset, neighborhood sensitivity-based sampling coreset, and maximum entropy coreset on full-body DIXON water and DIXON fat MRI images. All three coresets produced 27x compression with excellent performance in localizing five anatomical landmarks: left knee, right trochanter, left kidney, spleen, and lung across both imaging environments. Maximum entropy coreset obtained the best performance of $11.97\pm 12.02$ average distance error, compared to the conventional lifelong learning framework's $19.24\pm 50.77$.
Multi-environment lifelong deep reinforcement learning for medical imaging
May 31, 2023Abstract:Deep reinforcement learning(DRL) is increasingly being explored in medical imaging. However, the environments for medical imaging tasks are constantly evolving in terms of imaging orientations, imaging sequences, and pathologies. To that end, we developed a Lifelong DRL framework, SERIL to continually learn new tasks in changing imaging environments without catastrophic forgetting. SERIL was developed using selective experience replay based lifelong learning technique for the localization of five anatomical landmarks in brain MRI on a sequence of twenty-four different imaging environments. The performance of SERIL, when compared to two baseline setups: MERT(multi-environment-best-case) and SERT(single-environment-worst-case) demonstrated excellent performance with an average distance of $9.90\pm7.35$ pixels from the desired landmark across all 120 tasks, compared to $10.29\pm9.07$ for MERT and $36.37\pm22.41$ for SERT($p<0.05$), demonstrating the excellent potential for continuously learning multiple tasks across dynamically changing imaging environments.
Asynchronous Decentralized Federated Lifelong Learning for Landmark Localization in Medical Imaging
Mar 12, 2023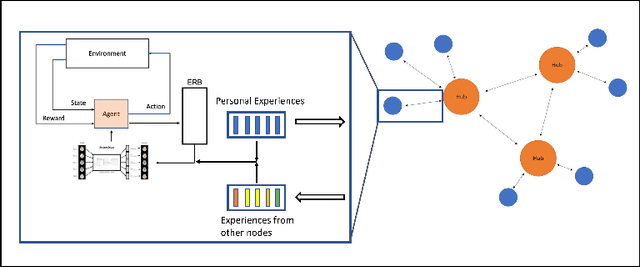
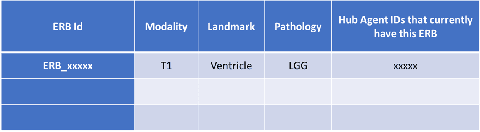
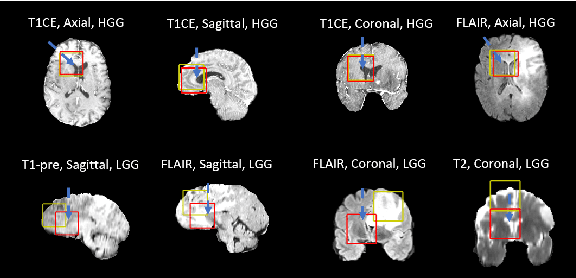
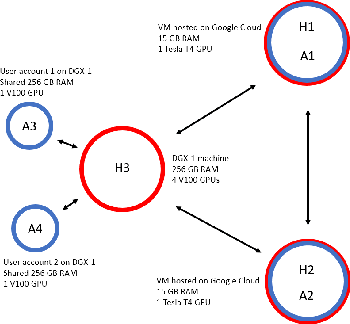
Abstract:Federated learning is a recent development in the machine learning area that allows a system of devices to train on one or more tasks without sharing their data to a single location or device. However, this framework still requires a centralized global model to consolidate individual models into one, and the devices train synchronously, which both can be potential bottlenecks for using federated learning. In this paper, we propose a novel method of asynchronous decentralized federated lifelong learning (ADFLL) method that inherits the merits of federated learning and can train on multiple tasks simultaneously without the need for a central node or synchronous training. Thus, overcoming the potential drawbacks of conventional federated learning. We demonstrate excellent performance on the brain tumor segmentation (BRATS) dataset for localizing the left ventricle on multiple image sequences and image orientation. Our framework allows agents to achieve the best performance with a mean distance error of 7.81, better than the conventional all-knowing agent's mean distance error of 11.78, and significantly (p=0.01) better than a conventional lifelong learning agent with a distance error of 15.17 after eight rounds of training. In addition, all ADFLL agents have comparable or better performance than a conventional LL agent. In conclusion, we developed an ADFLL framework with excellent performance and speed-up compared to conventional RL agents.
Selective experience replay compression using coresets for lifelong deep reinforcement learning in medical imaging
Feb 25, 2023Abstract:Selective experience replay is a popular strategy for integrating lifelong learning with deep reinforcement learning. Selective experience replay aims to recount selected experiences from previous tasks to avoid catastrophic forgetting. Furthermore, selective experience replay based techniques are model agnostic and allow experiences to be shared across different models. However, storing experiences from all previous tasks make lifelong learning using selective experience replay computationally very expensive and impractical as the number of tasks increase. To that end, we propose a reward distribution-preserving coreset compression technique for compressing experience replay buffers stored for selective experience replay. We evaluated the coreset compression technique on the brain tumor segmentation (BRATS) dataset for the task of ventricle localization and on the whole-body MRI for localization of left knee cap, left kidney, right trochanter, left lung, and spleen. The coreset lifelong learning models trained on a sequence of 10 different brain MR imaging environments demonstrated excellent performance localizing the ventricle with a mean pixel error distance of 12.93 for the compression ratio of 10x. In comparison, the conventional lifelong learning model localized the ventricle with a mean pixel distance of 10.87. Similarly, the coreset lifelong learning models trained on whole-body MRI demonstrated no significant difference (p=0.28) between the 10x compressed coreset lifelong learning models and conventional lifelong learning models for all the landmarks. The mean pixel distance for the 10x compressed models across all the landmarks was 25.30, compared to 19.24 for the conventional lifelong learning models. Our results demonstrate that the potential of the coreset-based ERB compression method for compressing experiences without a significant drop in performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge