Adway Kanhere
Improving Multi-Center Generalizability of GAN-Based Fat Suppression using Federated Learning
Apr 10, 2024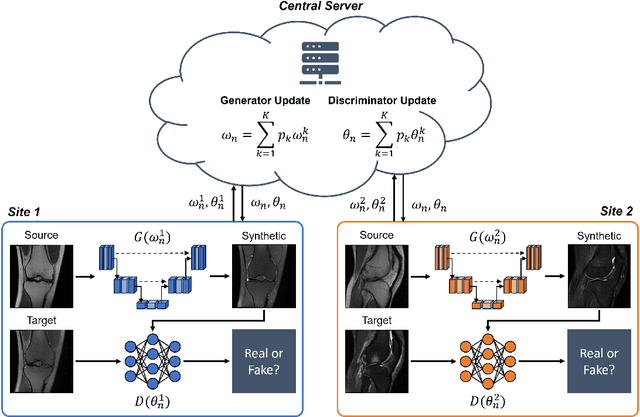
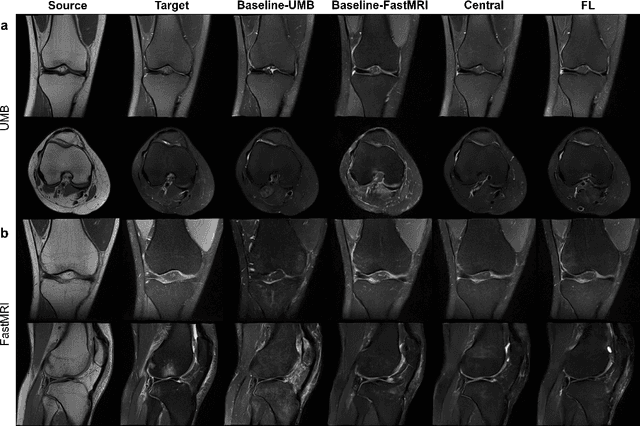
Abstract:Generative Adversarial Network (GAN)-based synthesis of fat suppressed (FS) MRIs from non-FS proton density sequences has the potential to accelerate acquisition of knee MRIs. However, GANs trained on single-site data have poor generalizability to external data. We show that federated learning can improve multi-center generalizability of GANs for synthesizing FS MRIs, while facilitating privacy-preserving multi-institutional collaborations.
Anytime, Anywhere, Anyone: Investigating the Feasibility of Segment Anything Model for Crowd-Sourcing Medical Image Annotations
Mar 22, 2024Abstract:Curating annotations for medical image segmentation is a labor-intensive and time-consuming task that requires domain expertise, resulting in "narrowly" focused deep learning (DL) models with limited translational utility. Recently, foundation models like the Segment Anything Model (SAM) have revolutionized semantic segmentation with exceptional zero-shot generalizability across various domains, including medical imaging, and hold a lot of promise for streamlining the annotation process. However, SAM has yet to be evaluated in a crowd-sourced setting to curate annotations for training 3D DL segmentation models. In this work, we explore the potential of SAM for crowd-sourcing "sparse" annotations from non-experts to generate "dense" segmentation masks for training 3D nnU-Net models, a state-of-the-art DL segmentation model. Our results indicate that while SAM-generated annotations exhibit high mean Dice scores compared to ground-truth annotations, nnU-Net models trained on SAM-generated annotations perform significantly worse than nnU-Net models trained on ground-truth annotations ($p<0.001$, all).
One Copy Is All You Need: Resource-Efficient Streaming of Medical Imaging Data at Scale
Jul 01, 2023Abstract:Large-scale medical imaging datasets have accelerated development of artificial intelligence tools for clinical decision support. However, the large size of these datasets is a bottleneck for users with limited storage and bandwidth. Many users may not even require such large datasets as AI models are often trained on lower resolution images. If users could directly download at their desired resolution, storage and bandwidth requirements would significantly decrease. However, it is impossible to anticipate every users' requirements and impractical to store the data at multiple resolutions. What if we could store images at a single resolution but send them at different ones? We propose MIST, an open-source framework to operationalize progressive resolution for streaming medical images at multiple resolutions from a single high-resolution copy. We demonstrate that MIST can dramatically reduce imaging infrastructure inefficiencies for hosting and streaming medical images by >90%, while maintaining diagnostic quality for deep learning applications.
High-Throughput AI Inference for Medical Image Classification and Segmentation using Intelligent Streaming
May 24, 2023Abstract:As the adoption of AI systems within the clinical setup grows, limitations in bandwidth could create communication bottlenecks when streaming imaging data, leading to delays in patient diagnosis and treatment. As such, healthcare providers and AI vendors will require greater computational infrastructure, therefore dramatically increasing costs. To that end, we developed intelligent streaming, a state-of-the-art framework to enable accelerated, cost-effective, bandwidth-optimized, and computationally efficient AI inference for clinical decision making at scale. For classification, intelligent streaming reduced the data transmission by 99.01% and decoding time by 98.58%, while increasing throughput by 27.43x. For segmentation, our framework reduced data transmission by 90.32%, decoding time by 90.26%, while increasing throughput by 4.20x. Our work demonstrates that intelligent streaming results in faster turnaround times, and reduced overall cost of data and transmission, without negatively impacting clinical decision making using AI systems.
Text2Cohort: Democratizing the NCI Imaging Data Commons with Natural Language Cohort Discovery
May 16, 2023Abstract:The Imaging Data Commons (IDC) is a cloud-based database that provides researchers with open access to cancer imaging data, with the goal of facilitating collaboration in medical imaging research. However, querying the IDC database for cohort discovery and access to imaging data has a significant learning curve for researchers due to its complex nature. We developed Text2Cohort, a large language model (LLM) based toolkit to facilitate user-friendly and intuitive natural language cohort discovery in the IDC. Text2Cohorts translates user input into IDC database queries using prompt engineering and autocorrection and returns the query's response to the user. Autocorrection resolves errors in queries by passing the errors back to the model for interpretation and correction. We evaluate Text2Cohort on 50 natural language user inputs ranging from information extraction to cohort discovery. The resulting queries and outputs were verified by two computer scientists to measure Text2Cohort's accuracy and F1 score. Text2Cohort successfully generated queries and their responses with an 88% accuracy and F1 score of 0.94. However, it failed to generate queries for 6/50 (12%) user inputs due to syntax and semantic errors. Our results indicate that Text2Cohort succeeded at generating queries with correct responses, but occasionally failed due to a lack of understanding of the data schema. Despite these shortcomings, Text2Cohort demonstrates the utility of LLMs to enable researchers to discover and curate cohorts using data hosted on IDC with high levels of accuracy using natural language in a more intuitive and user-friendly way.
Optimizing Federated Learning for Medical Image Classification on Distributed Non-iid Datasets with Partial Labels
Mar 10, 2023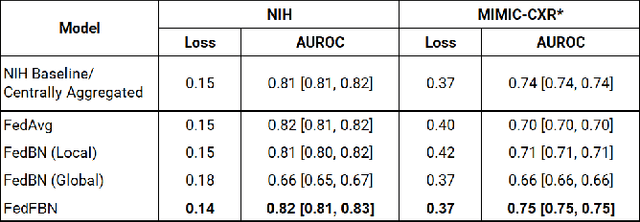
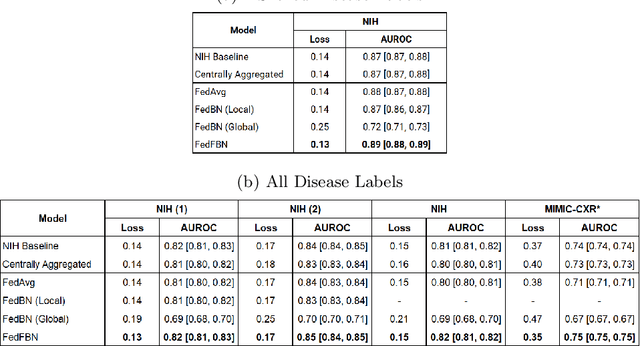
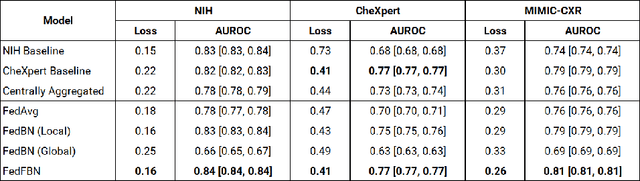
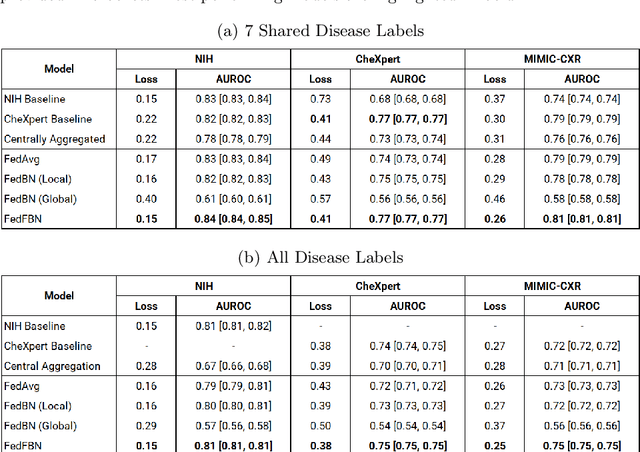
Abstract:Numerous large-scale chest x-ray datasets have spearheaded expert-level detection of abnormalities using deep learning. However, these datasets focus on detecting a subset of disease labels that could be present, thus making them distributed and non-iid with partial labels. Recent literature has indicated the impact of batch normalization layers on the convergence of federated learning due to domain shift associated with non-iid data with partial labels. To that end, we propose FedFBN, a federated learning framework that draws inspiration from transfer learning by using pretrained networks as the model backend and freezing the batch normalization layers throughout the training process. We evaluate FedFBN with current FL strategies using synthetic iid toy datasets and large-scale non-iid datasets across scenarios with partial and complete labels. Our results demonstrate that FedFBN outperforms current aggregation strategies for training global models using distributed and non-iid data with partial labels.
Surgical Aggregation: A Federated Learning Framework for Harmonizing Distributed Datasets with Diverse Tasks
Jan 17, 2023Abstract:AI-assisted characterization of chest x-rays (CXR) has the potential to provide substantial benefits across many clinical applications. Many large-scale public CXR datasets have been curated for detection of abnormalities using deep learning. However, each of these datasets focus on detecting a subset of disease labels that could be present in a CXR, thus limiting their clinical utility. Furthermore, the distributed nature of these datasets, along with data sharing regulations, make it difficult to share and create a complete representation of disease labels. We propose surgical aggregation, a federated learning framework for aggregating knowledge from distributed datasets with different disease labels into a 'global' deep learning model. We randomly divided the NIH Chest X-Ray 14 dataset into training (70%), validation (10%), and test (20%) splits with no patient overlap and conducted two experiments. In the first experiment, we pruned the disease labels to create two 'toy' datasets containing 11 and 8 labels respectively with 4 overlapping labels. For the second experiment, we pruned the disease labels to create two disjoint 'toy' datasets with 7 labels each. We observed that the surgically aggregated 'global' model resulted in excellent performance across both experiments when compared to a 'baseline' model trained on complete disease labels. The overlapping and disjoint experiments had an AUROC of 0.87 and 0.86 respectively, compared to the baseline AUROC of 0.87. We used surgical aggregation to harmonize the NIH Chest X-Ray 14 and CheXpert datasets into a 'global' model with an AUROC of 0.85 and 0.83 respectively. Our results show that surgical aggregation could be used to develop clinically useful deep learning models by aggregating knowledge from distributed datasets with diverse tasks, a step forward towards bridging the gap from bench to bedside.
From Competition to Collaboration: Making Toy Datasets on Kaggle Clinically Useful for Chest X-Ray Diagnosis Using Federated Learning
Nov 11, 2022



Abstract:Chest X-ray (CXR) datasets hosted on Kaggle, though useful from a data science competition standpoint, have limited utility in clinical use because of their narrow focus on diagnosing one specific disease. In real-world clinical use, multiple diseases need to be considered since they can co-exist in the same patient. In this work, we demonstrate how federated learning (FL) can be used to make these toy CXR datasets from Kaggle clinically useful. Specifically, we train a single FL classification model (`global`) using two separate CXR datasets -- one annotated for presence of pneumonia and the other for presence of pneumothorax (two common and life-threatening conditions) -- capable of diagnosing both. We compare the performance of the global FL model with models trained separately on both datasets (`baseline`) for two different model architectures. On a standard, naive 3-layer CNN architecture, the global FL model achieved AUROC of 0.84 and 0.81 for pneumonia and pneumothorax, respectively, compared to 0.85 and 0.82, respectively, for both baseline models (p>0.05). Similarly, on a pretrained DenseNet121 architecture, the global FL model achieved AUROC of 0.88 and 0.91 for pneumonia and pneumothorax, respectively, compared to 0.89 and 0.91, respectively, for both baseline models (p>0.05). Our results suggest that FL can be used to create global `meta` models to make toy datasets from Kaggle clinically useful, a step forward towards bridging the gap from bench to bedside.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge