Andrew Chan
Anytime, Anywhere, Anyone: Investigating the Feasibility of Segment Anything Model for Crowd-Sourcing Medical Image Annotations
Mar 22, 2024Abstract:Curating annotations for medical image segmentation is a labor-intensive and time-consuming task that requires domain expertise, resulting in "narrowly" focused deep learning (DL) models with limited translational utility. Recently, foundation models like the Segment Anything Model (SAM) have revolutionized semantic segmentation with exceptional zero-shot generalizability across various domains, including medical imaging, and hold a lot of promise for streamlining the annotation process. However, SAM has yet to be evaluated in a crowd-sourced setting to curate annotations for training 3D DL segmentation models. In this work, we explore the potential of SAM for crowd-sourcing "sparse" annotations from non-experts to generate "dense" segmentation masks for training 3D nnU-Net models, a state-of-the-art DL segmentation model. Our results indicate that while SAM-generated annotations exhibit high mean Dice scores compared to ground-truth annotations, nnU-Net models trained on SAM-generated annotations perform significantly worse than nnU-Net models trained on ground-truth annotations ($p<0.001$, all).
Hidden in Plain Sight: Undetectable Adversarial Bias Attacks on Vulnerable Patient Populations
Feb 08, 2024Abstract:The proliferation of artificial intelligence (AI) in radiology has shed light on the risk of deep learning (DL) models exacerbating clinical biases towards vulnerable patient populations. While prior literature has focused on quantifying biases exhibited by trained DL models, demographically targeted adversarial bias attacks on DL models and its implication in the clinical environment remains an underexplored field of research in medical imaging. In this work, we demonstrate that demographically targeted label poisoning attacks can introduce adversarial underdiagnosis bias in DL models and degrade performance on underrepresented groups without impacting overall model performance. Moreover, our results across multiple performance metrics and demographic groups like sex, age, and their intersectional subgroups indicate that a group's vulnerability to undetectable adversarial bias attacks is directly correlated with its representation in the model's training data.
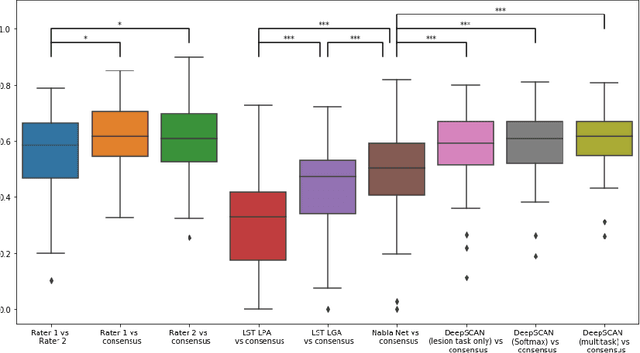
Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence
Apr 05, 2019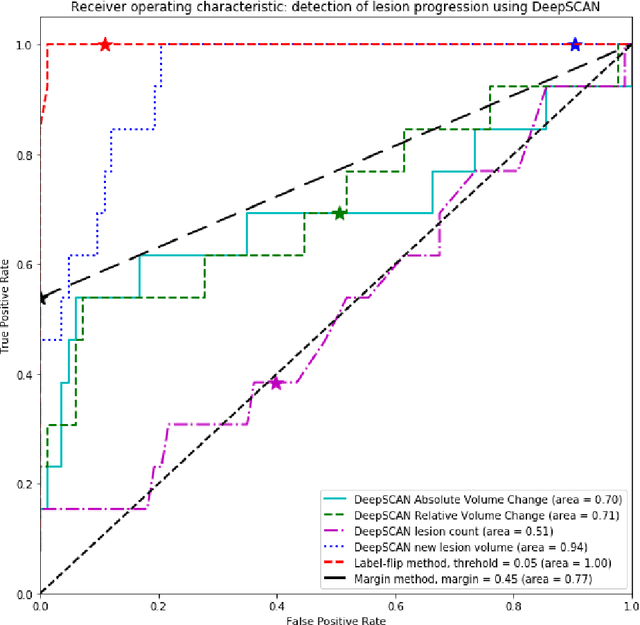
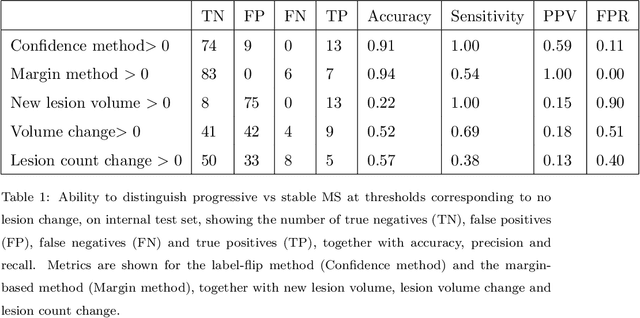
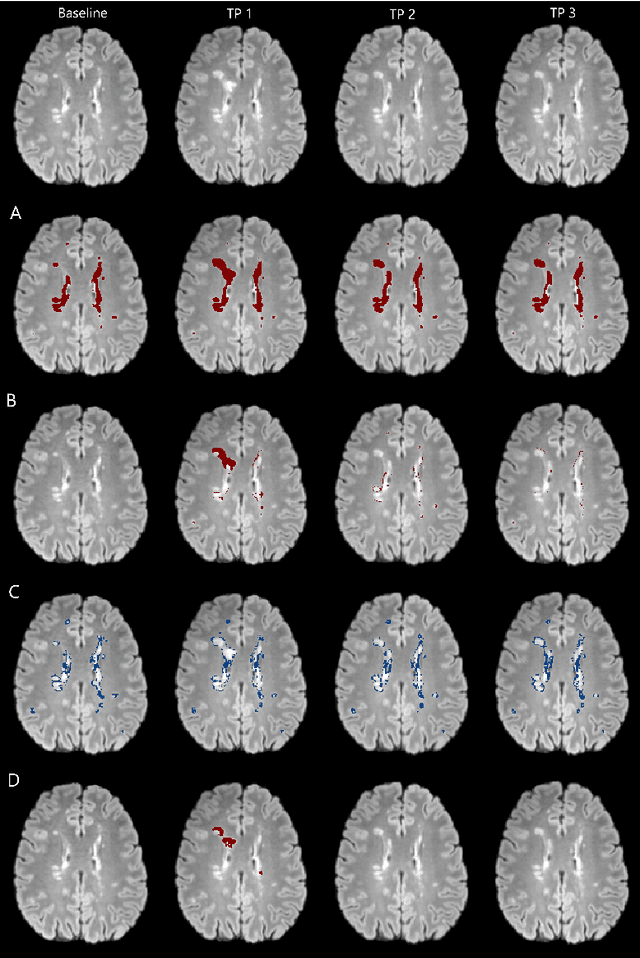
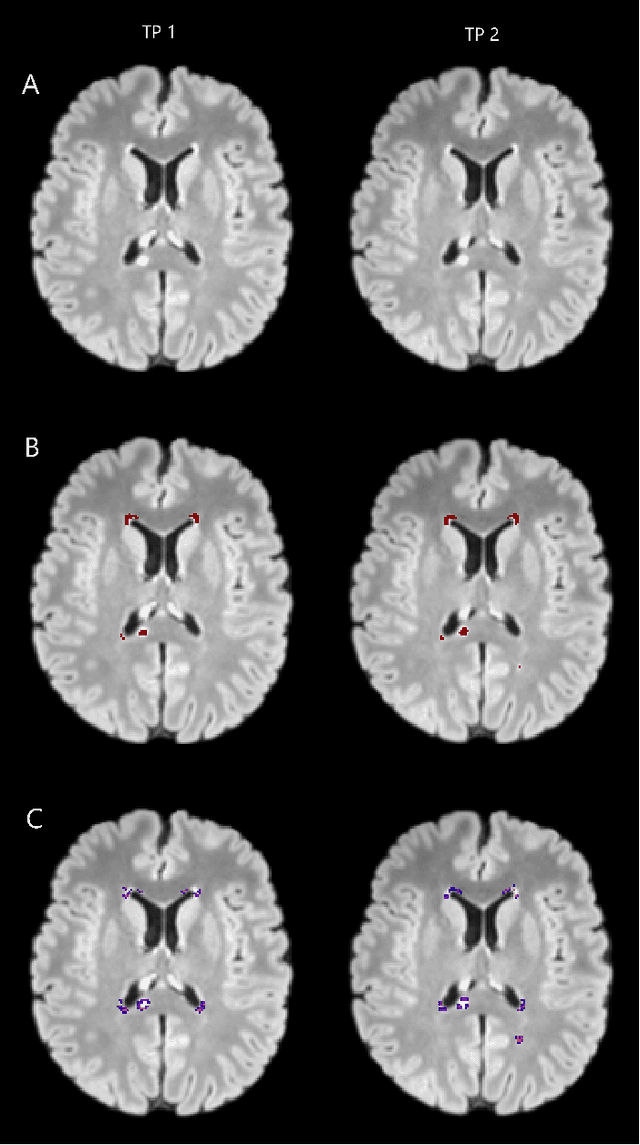
Abstract:The detection of new or enlarged white-matter lesions in multiple sclerosis is a vital task in the monitoring of patients undergoing disease-modifying treatment for multiple sclerosis. However, the definition of 'new or enlarged' is not fixed, and it is known that lesion-counting is highly subjective, with high degree of inter- and intra-rater variability. Automated methods for lesion quantification hold the potential to make the detection of new and enlarged lesions consistent and repeatable. However, the majority of lesion segmentation algorithms are not evaluated for their ability to separate progressive from stable patients, despite this being a pressing clinical use-case. In this paper we show that change in volumetric measurements of lesion load alone is not a good method for performing this separation, even for highly performing segmentation methods. Instead, we propose a method for identifying lesion changes of high certainty, and establish on a dataset of longitudinal multiple sclerosis cases that this method is able to separate progressive from stable timepoints with a very high level of discrimination (AUC = 0.99), while changes in lesion volume are much less able to perform this separation (AUC = 0.71). Validation of the method on a second external dataset confirms that the method is able to generalize beyond the setting in which it was trained, achieving an accuracy of 83% in separating stable and progressive timepoints. Both lesion volume and count have previously been shown to be strong predictors of disease course across a population. However, we demonstrate that for individual patients, changes in these measures are not an adequate means of establishing no evidence of disease activity. Meanwhile, directly detecting tissue which changes, with high confidence, from non-lesion to lesion is a feasible methodology for identifying radiologically active patients.
Simultaneous lesion and neuroanatomy segmentation in Multiple Sclerosis using deep neural networks
Jan 22, 2019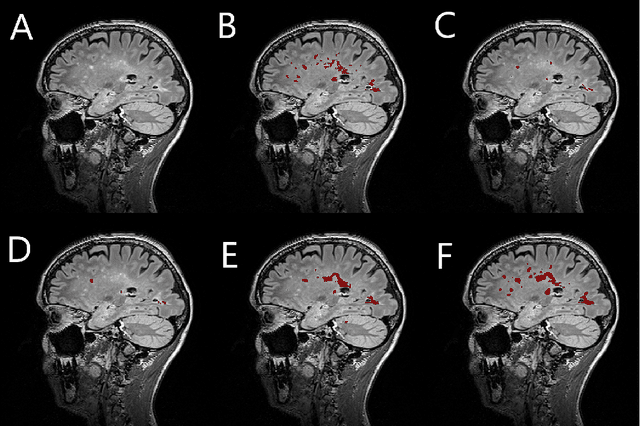
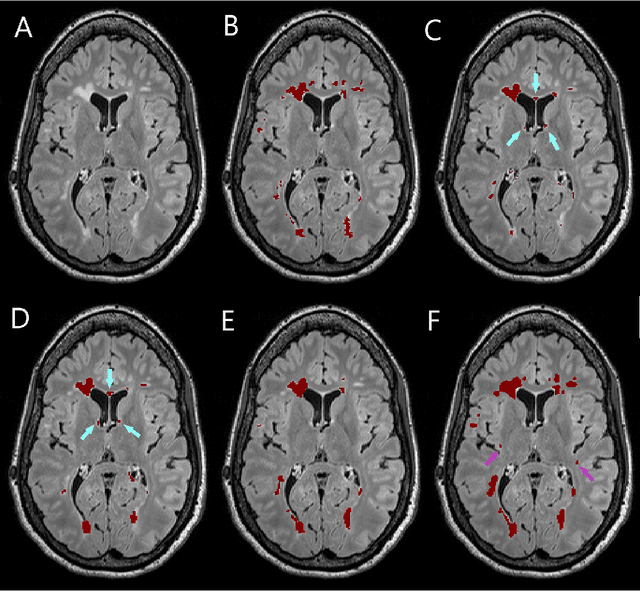
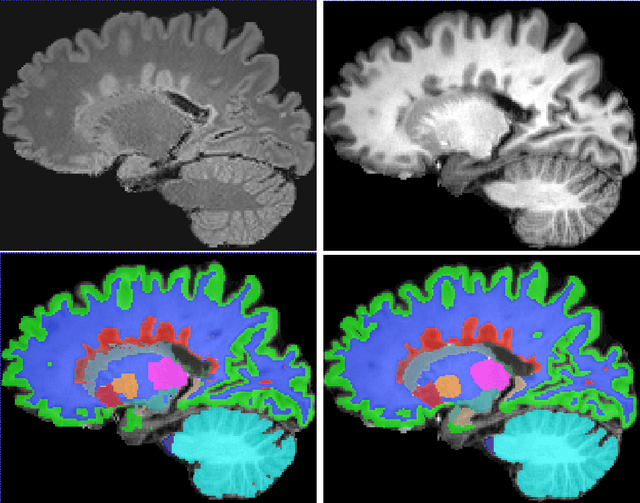
Abstract:Segmentation of both white matter lesions and deep grey matter structures is an important task in the quantification of magnetic resonance imaging in multiple sclerosis. Typically these tasks are performed separately: in this paper we present a single CNN-based segmentation solution for providing fast, reliable segmentations of multimodal MR imagies into lesion classes and healthy-appearing grey- and white-matter structures. We show substantial, statistically significant improvements in both Dice coefficient and in lesion-wise specificity and sensitivity, compared to previous approaches, and agreement with individual human raters in the range of human inter-rater variability. The method is trained on data gathered from a single centre: nonetheless, it performs well on data from centres, scanners and field-strengths not represented in the training dataset. A retrospective study found that the classifier successfully identified lesions missed by the human raters. Lesion labels were provided by human raters, while weak labels for other brain structures (including CSF, cortical grey matter, cortical white matter, cerebellum, amygdala, hippocampus, subcortical GM structures and choroid plexus) were provided by Freesurfer 5.3. The segmentations of these structures compared well, not only with Freesurfer 5.3, but also with FSL-First and Freesurfer 6.1.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge