Craig Jones
RTA-Former: Reverse Transformer Attention for Polyp Segmentation
Jan 22, 2024Abstract:Polyp segmentation is a key aspect of colorectal cancer prevention, enabling early detection and guiding subsequent treatments. Intelligent diagnostic tools, including deep learning solutions, are widely explored to streamline and potentially automate this process. However, even with many powerful network architectures, there still comes the problem of producing accurate edge segmentation. In this paper, we introduce a novel network, namely RTA-Former, that employs a transformer model as the encoder backbone and innovatively adapts Reverse Attention (RA) with a transformer stage in the decoder for enhanced edge segmentation. The results of the experiments illustrate that RTA-Former achieves state-of-the-art (SOTA) performance in five polyp segmentation datasets. The strong capability of RTA-Former holds promise in improving the accuracy of Transformer-based polyp segmentation, potentially leading to better clinical decisions and patient outcomes. Our code will be publicly available on GitHub.
Evidential Uncertainty Quantification: A Variance-Based Perspective
Nov 19, 2023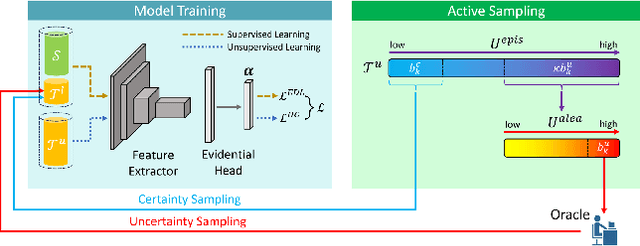
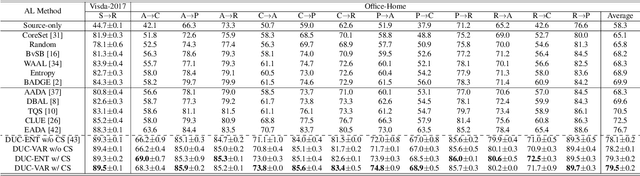
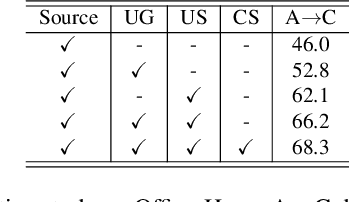
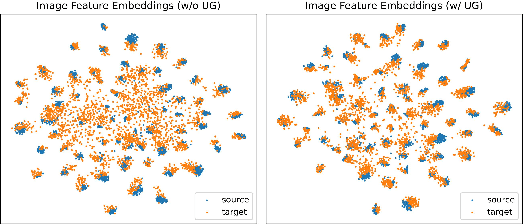
Abstract:Uncertainty quantification of deep neural networks has become an active field of research and plays a crucial role in various downstream tasks such as active learning. Recent advances in evidential deep learning shed light on the direct quantification of aleatoric and epistemic uncertainties with a single forward pass of the model. Most traditional approaches adopt an entropy-based method to derive evidential uncertainty in classification, quantifying uncertainty at the sample level. However, the variance-based method that has been widely applied in regression problems is seldom used in the classification setting. In this work, we adapt the variance-based approach from regression to classification, quantifying classification uncertainty at the class level. The variance decomposition technique in regression is extended to class covariance decomposition in classification based on the law of total covariance, and the class correlation is also derived from the covariance. Experiments on cross-domain datasets are conducted to illustrate that the variance-based approach not only results in similar accuracy as the entropy-based one in active domain adaptation but also brings information about class-wise uncertainties as well as between-class correlations. The code is available at https://github.com/KerryDRX/EvidentialADA. This alternative means of evidential uncertainty quantification will give researchers more options when class uncertainties and correlations are important in their applications.
Automated Artifact Detection in Ultra-widefield Fundus Photography of Patients with Sickle Cell Disease
Jul 11, 2023



Abstract:Importance: Ultra-widefield fundus photography (UWF-FP) has shown utility in sickle cell retinopathy screening; however, image artifact may diminish quality and gradeability of images. Objective: To create an automated algorithm for UWF-FP artifact classification. Design: A neural network based automated artifact detection algorithm was designed to identify commonly encountered UWF-FP artifacts in a cross section of patient UWF-FP. A pre-trained ResNet-50 neural network was trained on a subset of the images and the classification accuracy, sensitivity, and specificity were quantified on the hold out test set. Setting: The study is based on patients from a tertiary care hospital site. Participants: There were 243 UWF-FP acquired from patients with sickle cell disease (SCD), and artifact labelling in the following categories was performed: Eyelash Present, Lower Eyelid Obstructing, Upper Eyelid Obstructing, Image Too Dark, Dark Artifact, and Image Not Centered. Results: Overall, the accuracy for each class was Eyelash Present at 83.7%, Lower Eyelid Obstructing at 83.7%, Upper Eyelid Obstructing at 98.0%, Image Too Dark at 77.6%, Dark Artifact at 93.9%, and Image Not Centered at 91.8%. Conclusions and Relevance: This automated algorithm shows promise in identifying common imaging artifacts on a subset of Optos UWF-FP in SCD patients. Further refinement is ongoing with the goal of improving efficiency of tele-retinal screening in sickle cell retinopathy (SCR) by providing a photographer real-time feedback as to the types of artifacts present, and the need for image re-acquisition. This algorithm also may have potential future applicability in other retinal diseases by improving quality and efficiency of image acquisition of UWF-FP.
Active Learning in Brain Tumor Segmentation with Uncertainty Sampling, Annotation Redundancy Restriction, and Data Initialization
Feb 05, 2023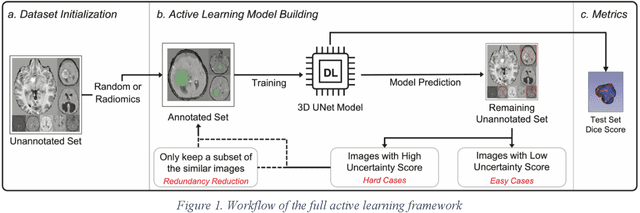
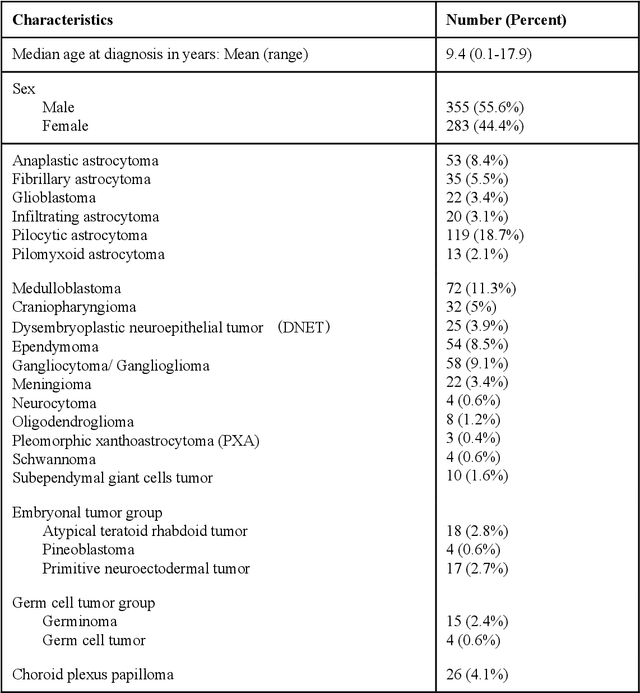
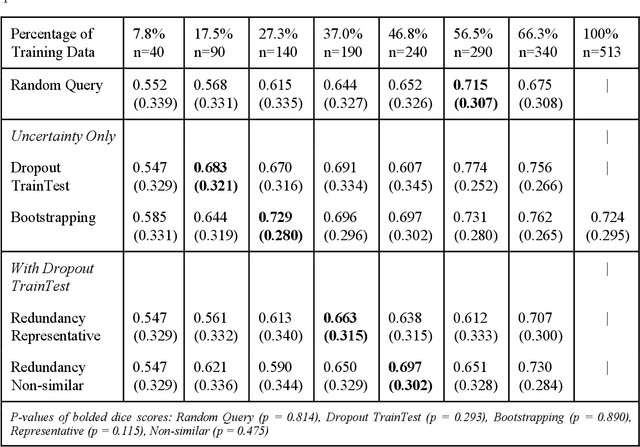
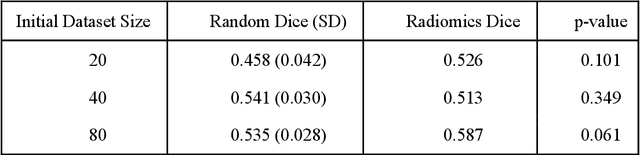
Abstract:Deep learning models have demonstrated great potential in medical 3D imaging, but their development is limited by the expensive, large volume of annotated data required. Active learning (AL) addresses this by training a model on a subset of the most informative data samples without compromising performance. We compared different AL strategies and propose a framework that minimizes the amount of data needed for state-of-the-art performance. 638 multi-institutional brain tumor MRI images were used to train a 3D U-net model and compare AL strategies. We investigated uncertainty sampling, annotation redundancy restriction, and initial dataset selection techniques. Uncertainty estimation techniques including Bayesian estimation with dropout, bootstrapping, and margins sampling were compared to random query. Strategies to avoid annotation redundancy by removing similar images within the to-be-annotated subset were considered as well. We determined the minimum amount of data necessary to achieve similar performance to the model trained on the full dataset ({\alpha} = 0.1). A variance-based selection strategy using radiomics to identify the initial training dataset is also proposed. Bayesian approximation with dropout at training and testing showed similar results to that of the full data model with less than 20% of the training data (p=0.293) compared to random query achieving similar performance at 56.5% of the training data (p=0.814). Annotation redundancy restriction techniques achieved state-of-the-art performance at approximately 40%-50% of the training data. Radiomics dataset initialization had higher Dice with initial dataset sizes of 20 and 80 images, but improvements were not significant. In conclusion, we investigated various AL strategies with dropout uncertainty estimation achieving state-of-the-art performance with the least annotated data.
Using Uncertainty in Deep Learning Reconstruction for Cone-Beam CT of the Brain
Aug 20, 2021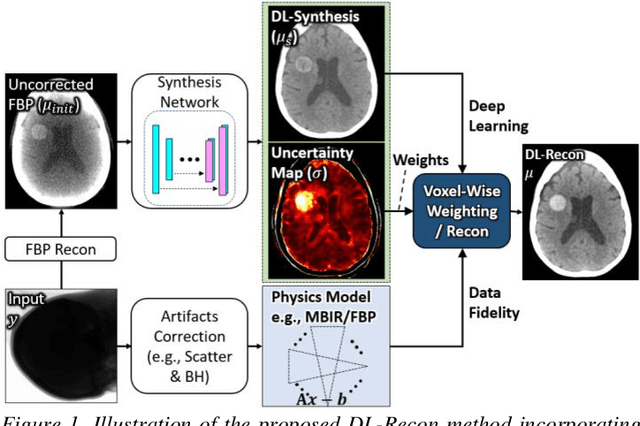
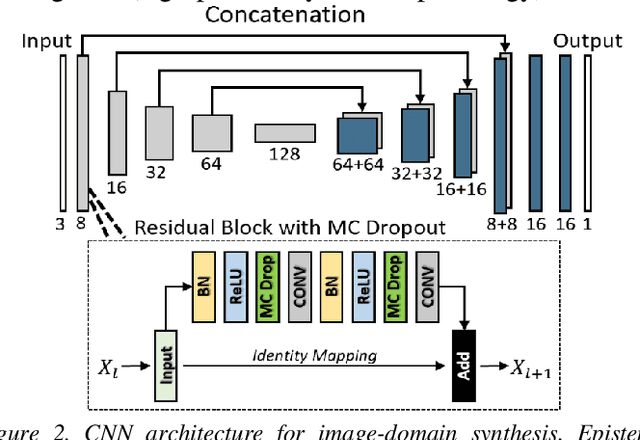
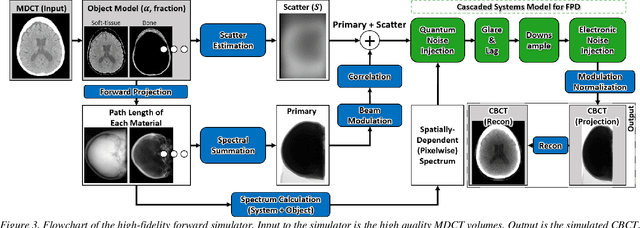
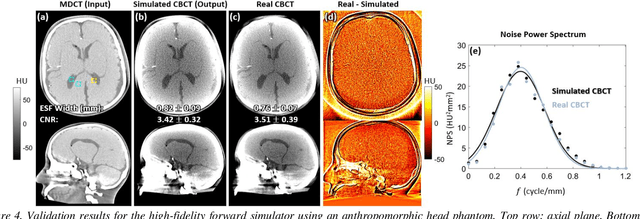
Abstract:Contrast resolution beyond the limits of conventional cone-beam CT (CBCT) systems is essential to high-quality imaging of the brain. We present a deep learning reconstruction method (dubbed DL-Recon) that integrates physically principled reconstruction models with DL-based image synthesis based on the statistical uncertainty in the synthesis image. A synthesis network was developed to generate a synthesized CBCT image (DL-Synthesis) from an uncorrected filtered back-projection (FBP) image. To improve generalizability (including accurate representation of lesions not seen in training), voxel-wise epistemic uncertainty of DL-Synthesis was computed using a Bayesian inference technique (Monte-Carlo dropout). In regions of high uncertainty, the DL-Recon method incorporates information from a physics-based reconstruction model and artifact-corrected projection data. Two forms of the DL-Recon method are proposed: (i) image-domain fusion of DL-Synthesis and FBP (DL-FBP) weighted by DL uncertainty; and (ii) a model-based iterative image reconstruction (MBIR) optimization using DL-Synthesis to compute a spatially varying regularization term based on DL uncertainty (DL-MBIR). The error in DL-Synthesis images was correlated with the uncertainty in the synthesis estimate. Compared to FBP and PWLS, the DL-Recon methods (both DL-FBP and DL-MBIR) showed ~50% reduction in noise (at matched spatial resolution) and ~40-70% improvement in image uniformity. Conventional DL-Synthesis alone exhibited ~10-60% under-estimation of lesion contrast and ~5-40% reduction in lesion segmentation accuracy (Dice coefficient) in simulated and real brain lesions, suggesting a lack of reliability / generalizability for structures unseen in the training data. DL-FBP and DL-MBIR improved the accuracy of reconstruction by directly incorporating information from the measurements in regions of high uncertainty.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge