Bram Stieltjes
A Multi-Centric Anthropomorphic 3D CT Phantom-Based Benchmark Dataset for Harmonization
Jul 02, 2025Abstract:Artificial intelligence (AI) has introduced numerous opportunities for human assistance and task automation in medicine. However, it suffers from poor generalization in the presence of shifts in the data distribution. In the context of AI-based computed tomography (CT) analysis, significant data distribution shifts can be caused by changes in scanner manufacturer, reconstruction technique or dose. AI harmonization techniques can address this problem by reducing distribution shifts caused by various acquisition settings. This paper presents an open-source benchmark dataset containing CT scans of an anthropomorphic phantom acquired with various scanners and settings, which purpose is to foster the development of AI harmonization techniques. Using a phantom allows fixing variations attributed to inter- and intra-patient variations. The dataset includes 1378 image series acquired with 13 scanners from 4 manufacturers across 8 institutions using a harmonized protocol as well as several acquisition doses. Additionally, we present a methodology, baseline results and open-source code to assess image- and feature-level stability and liver tissue classification, promoting the development of AI harmonization strategies.
Clinnova Federated Learning Proof of Concept: Key Takeaways from a Cross-border Collaboration
Oct 03, 2024



Abstract:Clinnova, a collaborative initiative involving France, Germany, Switzerland, and Luxembourg, is dedicated to unlocking the power of precision medicine through data federation, standardization, and interoperability. This European Greater Region initiative seeks to create an interoperable European standard using artificial intelligence (AI) and data science to enhance healthcare outcomes and efficiency. Key components include multidisciplinary research centers, a federated biobanking strategy, a digital health innovation platform, and a federated AI strategy. It targets inflammatory bowel disease, rheumatoid diseases, and multiple sclerosis (MS), emphasizing data quality to develop AI algorithms for personalized treatment and translational research. The IHU Strasbourg (Institute of Minimal-invasive Surgery) has the lead in this initiative to develop the federated learning (FL) proof of concept (POC) that will serve as a foundation for advancing AI in healthcare. At its core, Clinnova-MS aims to enhance MS patient care by using FL to develop more accurate models that detect disease progression, guide interventions, and validate digital biomarkers across multiple sites. This technical report presents insights and key takeaways from the first cross-border federated POC on MS segmentation of MRI images within the Clinnova framework. While our work marks a significant milestone in advancing MS segmentation through cross-border collaboration, it also underscores the importance of addressing technical, logistical, and ethical considerations to realize the full potential of FL in healthcare settings.
DALSA: Domain Adaptation for Supervised Learning From Sparsely Annotated MR Images
Mar 12, 2024
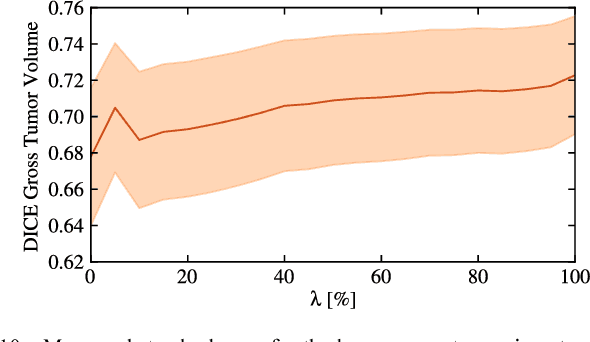

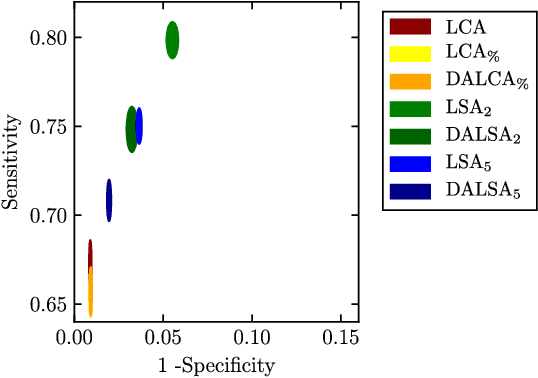
Abstract:We propose a new method that employs transfer learning techniques to effectively correct sampling selection errors introduced by sparse annotations during supervised learning for automated tumor segmentation. The practicality of current learning-based automated tissue classification approaches is severely impeded by their dependency on manually segmented training databases that need to be recreated for each scenario of application, site, or acquisition setup. The comprehensive annotation of reference datasets can be highly labor-intensive, complex, and error-prone. The proposed method derives high-quality classifiers for the different tissue classes from sparse and unambiguous annotations and employs domain adaptation techniques for effectively correcting sampling selection errors introduced by the sparse sampling. The new approach is validated on labeled, multi-modal MR images of 19 patients with malignant gliomas and by comparative analysis on the BraTS 2013 challenge data sets. Compared to training on fully labeled data, we reduced the time for labeling and training by a factor greater than 70 and 180 respectively without sacrificing accuracy. This dramatically eases the establishment and constant extension of large annotated databases in various scenarios and imaging setups and thus represents an important step towards practical applicability of learning-based approaches in tissue classification.
Common Limitations of Image Processing Metrics: A Picture Story
Apr 13, 2021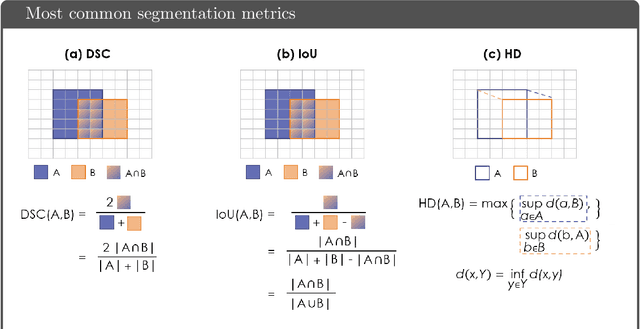
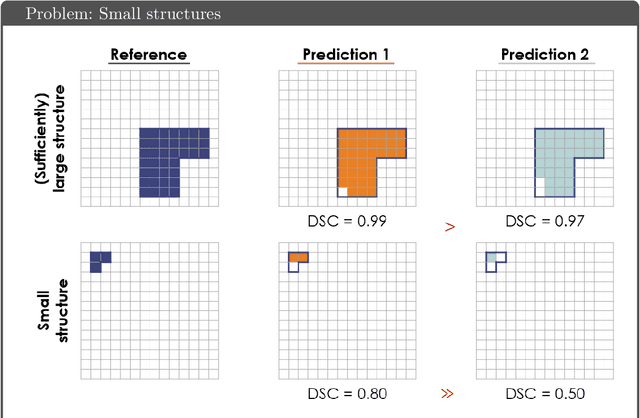
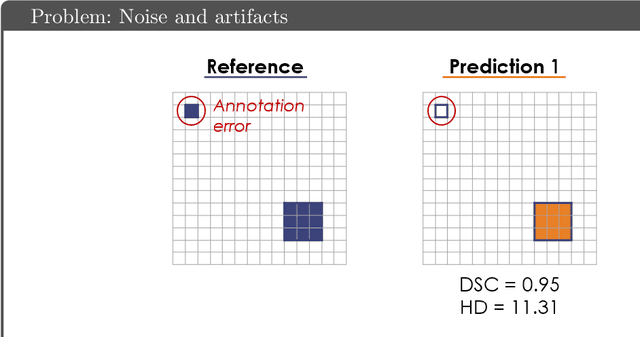
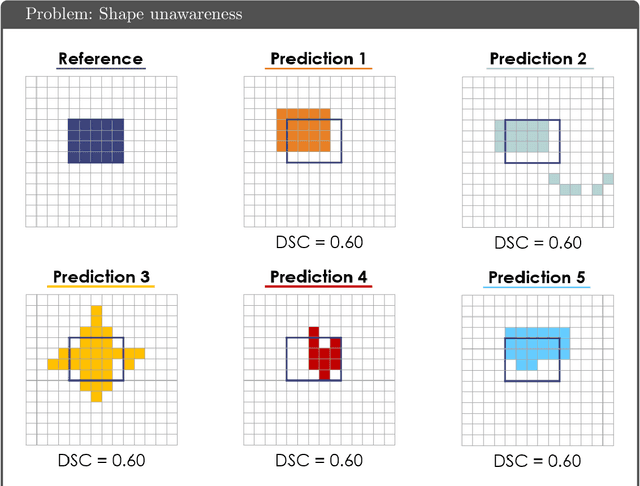
Abstract:While the importance of automatic image analysis is increasing at an enormous pace, recent meta-research revealed major flaws with respect to algorithm validation. Specifically, performance metrics are key for objective, transparent and comparative performance assessment, but relatively little attention has been given to the practical pitfalls when using specific metrics for a given image analysis task. A common mission of several international initiatives is therefore to provide researchers with guidelines and tools to choose the performance metrics in a problem-aware manner. This dynamically updated document has the purpose to illustrate important limitations of performance metrics commonly applied in the field of image analysis. The current version is based on a Delphi process on metrics conducted by an international consortium of image analysis experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge