Barbara Bobek-Billewicz
DALSA: Domain Adaptation for Supervised Learning From Sparsely Annotated MR Images
Mar 12, 2024
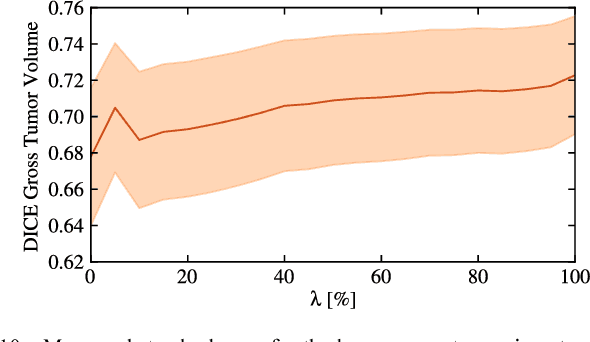

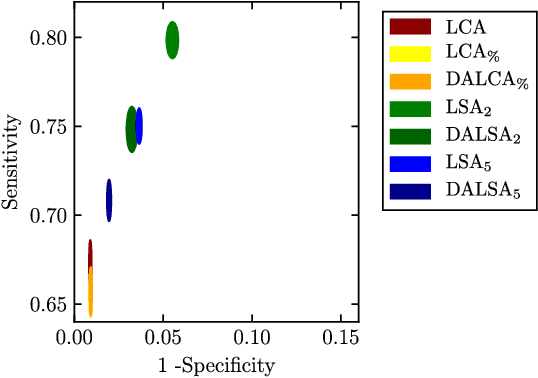
Abstract:We propose a new method that employs transfer learning techniques to effectively correct sampling selection errors introduced by sparse annotations during supervised learning for automated tumor segmentation. The practicality of current learning-based automated tissue classification approaches is severely impeded by their dependency on manually segmented training databases that need to be recreated for each scenario of application, site, or acquisition setup. The comprehensive annotation of reference datasets can be highly labor-intensive, complex, and error-prone. The proposed method derives high-quality classifiers for the different tissue classes from sparse and unambiguous annotations and employs domain adaptation techniques for effectively correcting sampling selection errors introduced by the sparse sampling. The new approach is validated on labeled, multi-modal MR images of 19 patients with malignant gliomas and by comparative analysis on the BraTS 2013 challenge data sets. Compared to training on fully labeled data, we reduced the time for labeling and training by a factor greater than 70 and 180 respectively without sacrificing accuracy. This dramatically eases the establishment and constant extension of large annotated databases in various scenarios and imaging setups and thus represents an important step towards practical applicability of learning-based approaches in tissue classification.
Fully-automated deep learning-powered system for DCE-MRI analysis of brain tumors
Jul 18, 2019



Abstract:Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) plays an important role in diagnosis and grading of brain tumor. Although manual DCE biomarker extraction algorithms boost the diagnostic yield of DCE-MRI by providing quantitative information on tumor prognosis and prediction, they are time-consuming and prone to human error. In this paper, we propose a fully-automated, end-to-end system for DCE-MRI analysis of brain tumors. Our deep learning-powered technique does not require any user interaction, it yields reproducible results, and it is rigorously validated against benchmark (BraTS'17 for tumor segmentation, and a test dataset released by the Quantitative Imaging Biomarkers Alliance for the contrast-concentration fitting) and clinical (44 low-grade glioma patients) data. Also, we introduce a cubic model of the vascular input function used for pharmacokinetic modeling which significantly decreases the fitting error when compared with the state of the art, alongside a real-time algorithm for determination of the vascular input region. An extensive experimental study, backed up with statistical tests, showed that our system delivers state-of-the-art results (in terms of segmentation accuracy and contrast-concentration fitting) while requiring less than 3 minutes to process an entire input DCE-MRI study using a single GPU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge