Bolin Li
Generative Multi-Focus Image Fusion
Dec 25, 2025



Abstract:Multi-focus image fusion aims to generate an all-in-focus image from a sequence of partially focused input images. Existing fusion algorithms generally assume that, for every spatial location in the scene, there is at least one input image in which that location is in focus. Furthermore, current fusion models often suffer from edge artifacts caused by uncertain focus estimation or hard-selection operations in complex real-world scenarios. To address these limitations, we propose a generative multi-focus image fusion framework, termed GMFF, which operates in two sequential stages. In the first stage, deterministic fusion is implemented using StackMFF V4, the latest version of the StackMFF series, and integrates the available focal plane information to produce an initial fused image. The second stage, generative restoration, is realized through IFControlNet, which leverages the generative capabilities of latent diffusion models to reconstruct content from missing focal planes, restore fine details, and eliminate edge artifacts. Each stage is independently developed and functions seamlessly in a cascaded manner. Extensive experiments demonstrate that GMFF achieves state-of-the-art fusion performance and exhibits significant potential for practical applications, particularly in scenarios involving complex multi-focal content. The implementation is publicly available at https://github.com/Xinzhe99/StackMFF-Series.
Periodic Bipedal Gait Learning Using Reward Composition Based on a Novel Gait Planner for Humanoid Robots
Jun 10, 2025Abstract:This paper presents a periodic bipedal gait learning method using reward composition, integrated with a real-time gait planner for humanoid robots. First, we introduce a novel gait planner that incorporates dynamics to design the desired joint trajectory. In the gait design process, the 3D robot model is decoupled into two 2D models, which are then approximated as hybrid inverted pendulums (H-LIP) for trajectory planning. The gait planner operates in parallel in real time within the robot's learning environment. Second, based on this gait planner, we design three effective reward functions within a reinforcement learning framework, forming a reward composition to achieve periodic bipedal gait. This reward composition reduces the robot's learning time and enhances locomotion performance. Finally, a gait design example and performance comparison are presented to demonstrate the effectiveness of the proposed method.
Boosting Private Domain Understanding of Efficient MLLMs: A Tuning-free, Adaptive, Universal Prompt Optimization Framework
Dec 27, 2024Abstract:Efficient multimodal large language models (EMLLMs), in contrast to multimodal large language models (MLLMs), reduce model size and computational costs and are often deployed on resource-constrained devices. However, due to data privacy concerns, existing open-source EMLLMs rarely have access to private domain-specific data during the pre-training process, making them difficult to directly apply in device-specific domains, such as certain business scenarios. To address this weakness, this paper focuses on the efficient adaptation of EMLLMs to private domains, specifically in two areas: 1) how to reduce data requirements, and 2) how to avoid parameter fine-tuning. Specifically, we propose a tun\textbf{\underline{I}}ng-free, a\textbf{\underline{D}}aptiv\textbf{\underline{E}}, univers\textbf{\underline{AL}} \textbf{\underline{Prompt}} Optimization Framework, abbreviated as \textit{\textbf{\ourmethod{}}} which consists of two stages: 1) Predefined Prompt, based on the reinforcement searching strategy, generate a prompt optimization strategy tree to acquire optimization priors; 2) Prompt Reflection initializes the prompt based on optimization priors, followed by self-reflection to further search and refine the prompt. By doing so, \ourmethod{} elegantly generates the ``ideal prompts'' for processing private domain-specific data. Note that our method requires no parameter fine-tuning and only a small amount of data to quickly adapt to the data distribution of private data. Extensive experiments across multiple tasks demonstrate that our proposed \ourmethod{} significantly improves both efficiency and performance compared to baselines.
LLaVA-MoD: Making LLaVA Tiny via MoE Knowledge Distillation
Aug 28, 2024Abstract:We introduce LLaVA-MoD, a novel framework designed to enable the efficient training of small-scale Multimodal Language Models (s-MLLM) by distilling knowledge from large-scale MLLM (l-MLLM). Our approach tackles two fundamental challenges in MLLM distillation. First, we optimize the network structure of s-MLLM by integrating a sparse Mixture of Experts (MoE) architecture into the language model, striking a balance between computational efficiency and model expressiveness. Second, we propose a progressive knowledge transfer strategy to ensure comprehensive knowledge migration. This strategy begins with mimic distillation, where we minimize the Kullback-Leibler (KL) divergence between output distributions to enable the student model to emulate the teacher network's understanding. Following this, we introduce preference distillation via Direct Preference Optimization (DPO), where the key lies in treating l-MLLM as the reference model. During this phase, the s-MLLM's ability to discriminate between superior and inferior examples is significantly enhanced beyond l-MLLM, leading to a better student that surpasses its teacher, particularly in hallucination benchmarks. Extensive experiments demonstrate that LLaVA-MoD outperforms existing models across various multimodal benchmarks while maintaining a minimal number of activated parameters and low computational costs. Remarkably, LLaVA-MoD, with only 2B activated parameters, surpasses Qwen-VL-Chat-7B by an average of 8.8% across benchmarks, using merely 0.3% of the training data and 23% trainable parameters. These results underscore LLaVA-MoD's ability to effectively distill comprehensive knowledge from its teacher model, paving the way for the development of more efficient MLLMs. The code will be available on: https://github.com/shufangxun/LLaVA-MoD.
CRT-Net: A Generalized and Scalable Framework for the Computer-Aided Diagnosis of Electrocardiogram Signals
May 28, 2021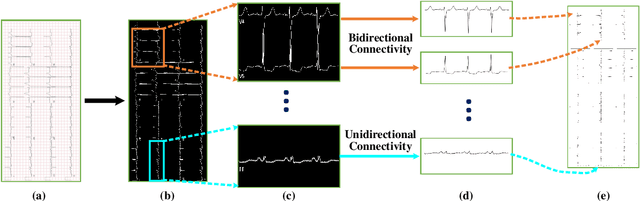
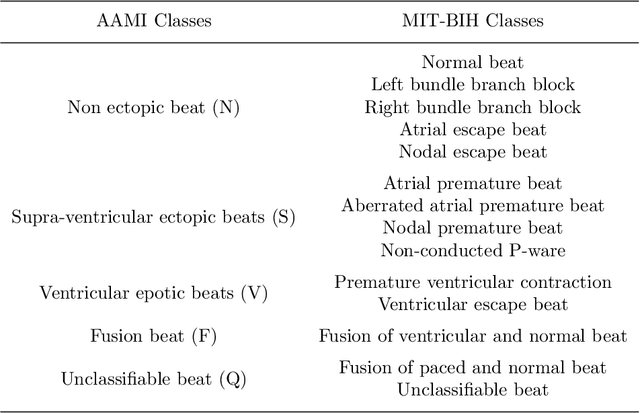
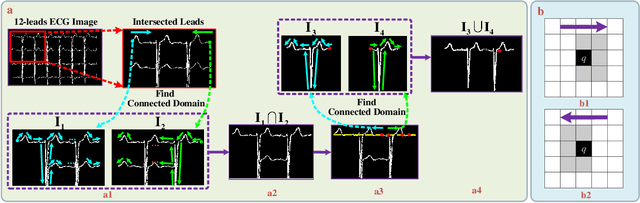
Abstract:Electrocardiogram (ECG) signals play critical roles in the clinical screening and diagnosis of many types of cardiovascular diseases. Despite deep neural networks that have been greatly facilitated computer-aided diagnosis (CAD) in many clinical tasks, the variability and complexity of ECG in the clinic still pose significant challenges in both diagnostic performance and clinical applications. In this paper, we develop a robust and scalable framework for the clinical recognition of ECG. Considering the fact that hospitals generally record ECG signals in the form of graphic waves of 2-D images, we first extract the graphic waves of 12-lead images into numerical 1-D ECG signals by a proposed bi-directional connectivity method. Subsequently, a novel deep neural network, namely CRT-Net, is designed for the fine-grained and comprehensive representation and recognition of 1-D ECG signals. The CRT-Net can well explore waveform features, morphological characteristics and time domain features of ECG by embedding convolution neural network(CNN), recurrent neural network(RNN), and transformer module in a scalable deep model, which is especially suitable in clinical scenarios with different lengths of ECG signals captured from different devices. The proposed framework is first evaluated on two widely investigated public repositories, demonstrating the superior performance of ECG recognition in comparison with state-of-the-art. Moreover, we validate the effectiveness of our proposed bi-directional connectivity and CRT-Net on clinical ECG images collected from the local hospital, including 258 patients with chronic kidney disease (CKD), 351 patients with Type-2 Diabetes (T2DM), and around 300 patients in the control group. In the experiments, our methods can achieve excellent performance in the recognition of these two types of disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge