Benteng Ma
Toward Robust Medical Fairness: Debiased Dual-Modal Alignment via Text-Guided Attribute-Disentangled Prompt Learning for Vision-Language Models
Aug 26, 2025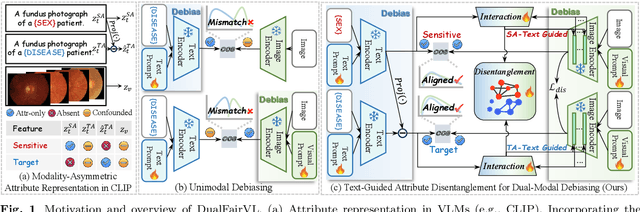
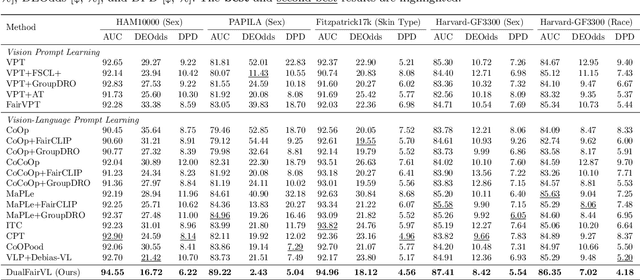
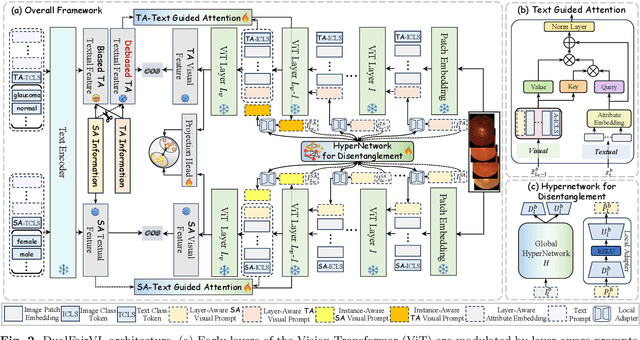
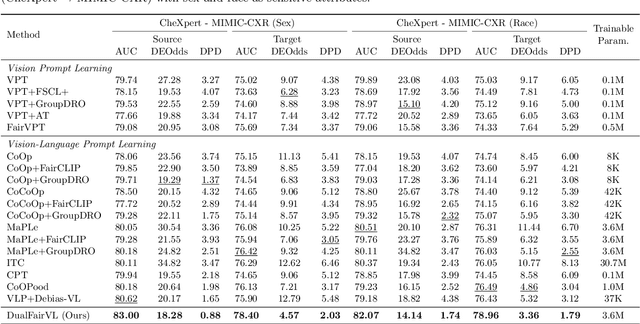
Abstract:Ensuring fairness across demographic groups in medical diagnosis is essential for equitable healthcare, particularly under distribution shifts caused by variations in imaging equipment and clinical practice. Vision-language models (VLMs) exhibit strong generalization, and text prompts encode identity attributes, enabling explicit identification and removal of sensitive directions. However, existing debiasing approaches typically address vision and text modalities independently, leaving residual cross-modal misalignment and fairness gaps. To address this challenge, we propose DualFairVL, a multimodal prompt-learning framework that jointly debiases and aligns cross-modal representations. DualFairVL employs a parallel dual-branch architecture that separates sensitive and target attributes, enabling disentangled yet aligned representations across modalities. Approximately orthogonal text anchors are constructed via linear projections, guiding cross-attention mechanisms to produce fused features. A hypernetwork further disentangles attribute-related information and generates instance-aware visual prompts, which encode dual-modal cues for fairness and robustness. Prototype-based regularization is applied in the visual branch to enforce separation of sensitive features and strengthen alignment with textual anchors. Extensive experiments on eight medical imaging datasets across four modalities show that DualFairVL achieves state-of-the-art fairness and accuracy under both in- and out-of-distribution settings, outperforming full fine-tuning and parameter-efficient baselines with only 3.6M trainable parameters. Code will be released upon publication.
Think Twice Before Selection: Federated Evidential Active Learning for Medical Image Analysis with Domain Shifts
Dec 05, 2023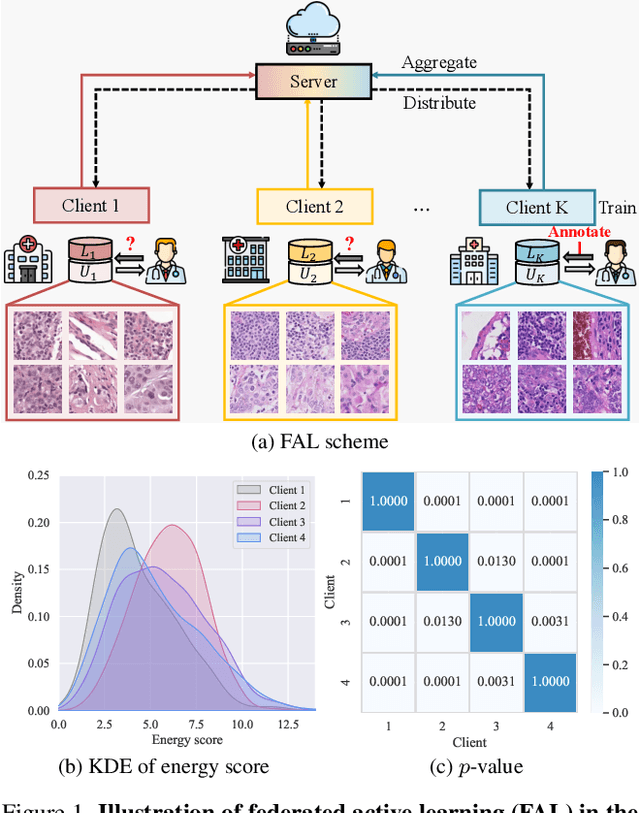
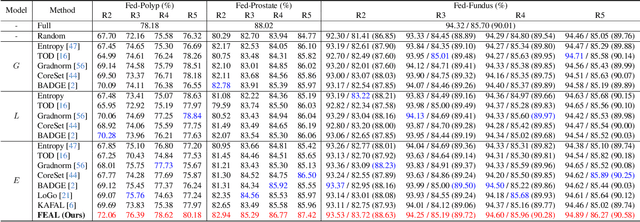
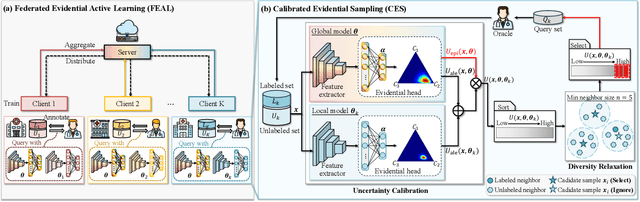
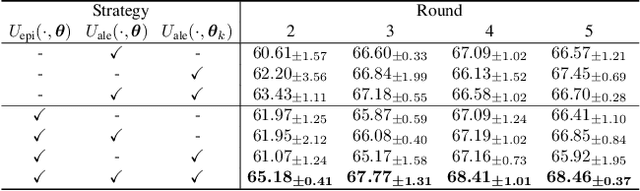
Abstract:Federated learning facilitates the collaborative learning of a global model across multiple distributed medical institutions without centralizing data. Nevertheless, the expensive cost of annotation on local clients remains an obstacle to effectively utilizing local data. To mitigate this issue, federated active learning methods suggest leveraging local and global model predictions to select a relatively small amount of informative local data for annotation. However, existing methods mainly focus on all local data sampled from the same domain, making them unreliable in realistic medical scenarios with domain shifts among different clients. In this paper, we make the first attempt to assess the informativeness of local data derived from diverse domains and propose a novel methodology termed Federated Evidential Active Learning (FEAL) to calibrate the data evaluation under domain shift. Specifically, we introduce a Dirichlet prior distribution in both local and global models to treat the prediction as a distribution over the probability simplex and capture both aleatoric and epistemic uncertainties by using the Dirichlet-based evidential model. Then we employ the epistemic uncertainty to calibrate the aleatoric uncertainty. Afterward, we design a diversity relaxation strategy to reduce data redundancy and maintain data diversity. Extensive experiments and analyses are conducted to show the superiority of FEAL over the state-of-the-art active learning methods and the efficiency of FEAL under the federated active learning framework.
Tackling the Incomplete Annotation Issue in Universal Lesion Detection Task By Exploratory Training
Sep 23, 2023Abstract:Universal lesion detection has great value for clinical practice as it aims to detect various types of lesions in multiple organs on medical images. Deep learning methods have shown promising results, but demanding large volumes of annotated data for training. However, annotating medical images is costly and requires specialized knowledge. The diverse forms and contrasts of objects in medical images make fully annotation even more challenging, resulting in incomplete annotations. Directly training ULD detectors on such datasets can yield suboptimal results. Pseudo-label-based methods examine the training data and mine unlabelled objects for retraining, which have shown to be effective to tackle this issue. Presently, top-performing methods rely on a dynamic label-mining mechanism, operating at the mini-batch level. However, the model's performance varies at different iterations, leading to inconsistencies in the quality of the mined labels and limits their performance enhancement. Inspired by the observation that deep models learn concepts with increasing complexity, we introduce an innovative exploratory training to assess the reliability of mined lesions over time. Specifically, we introduce a teacher-student detection model as basis, where the teacher's predictions are combined with incomplete annotations to train the student. Additionally, we design a prediction bank to record high-confidence predictions. Each sample is trained several times, allowing us to get a sequence of records for each sample. If a prediction consistently appears in the record sequence, it is likely to be a true object, otherwise it may just a noise. This serves as a crucial criterion for selecting reliable mined lesions for retraining. Our experimental results substantiate that the proposed framework surpasses state-of-the-art methods on two medical image datasets, demonstrating its superior performance.
Analyzing Deep Learning Based Brain Tumor Segmentation with Missing MRI Modalities
Aug 06, 2022Abstract:This technical report presents a comparative analysis of existing deep learning (DL) based approaches for brain tumor segmentation with missing MRI modalities. Approaches evaluated include the Adversarial Co-training Network (ACN) and a combination of mmGAN and DeepMedic. A more stable and easy-to-use version of mmGAN is also open-sourced at a GitHub repository. Using the BraTS2018 dataset, this work demonstrates that the state-of-the-art ACN performs better especially when T1c is missing. While a simple combination of mmGAN and DeepMedic also shows strong potentials when only one MRI modality is missing. Additionally, this work initiated discussions with future research directions for brain tumor segmentation with missing MRI modalities.
FIBA: Frequency-Injection based Backdoor Attack in Medical Image Analysis
Dec 02, 2021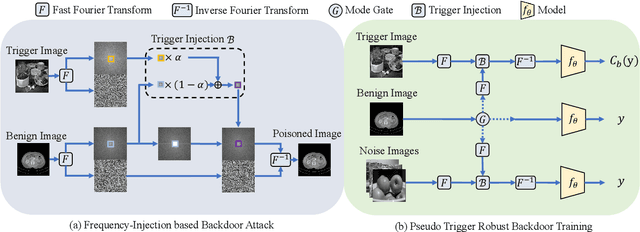
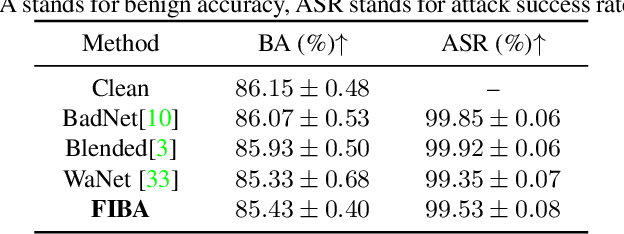
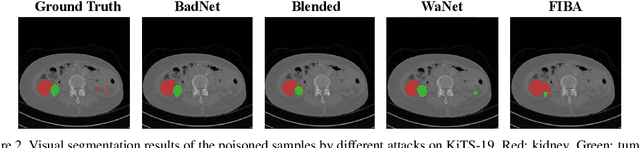
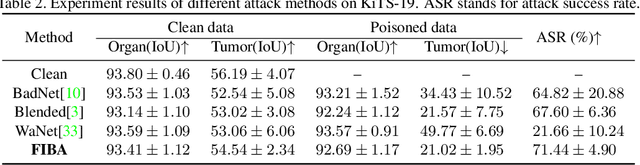
Abstract:In recent years, the security of AI systems has drawn increasing research attention, especially in the medical imaging realm. To develop a secure medical image analysis (MIA) system, it is a must to study possible backdoor attacks (BAs), which can embed hidden malicious behaviors into the system. However, designing a unified BA method that can be applied to various MIA systems is challenging due to the diversity of imaging modalities (e.g., X-Ray, CT, and MRI) and analysis tasks (e.g., classification, detection, and segmentation). Most existing BA methods are designed to attack natural image classification models, which apply spatial triggers to training images and inevitably corrupt the semantics of poisoned pixels, leading to the failures of attacking dense prediction models. To address this issue, we propose a novel Frequency-Injection based Backdoor Attack method (FIBA) that is capable of delivering attacks in various MIA tasks. Specifically, FIBA leverages a trigger function in the frequency domain that can inject the low-frequency information of a trigger image into the poisoned image by linearly combining the spectral amplitude of both images. Since it preserves the semantics of the poisoned image pixels, FIBA can perform attacks on both classification and dense prediction models. Experiments on three benchmarks in MIA (i.e., ISIC-2019 for skin lesion classification, KiTS-19 for kidney tumor segmentation, and EAD-2019 for endoscopic artifact detection), validate the effectiveness of FIBA and its superiority over state-of-the-art methods in attacking MIA models as well as bypassing backdoor defense. The code will be available at https://github.com/HazardFY/FIBA.
Inter-layer Transition in Neural Architecture Search
Nov 30, 2020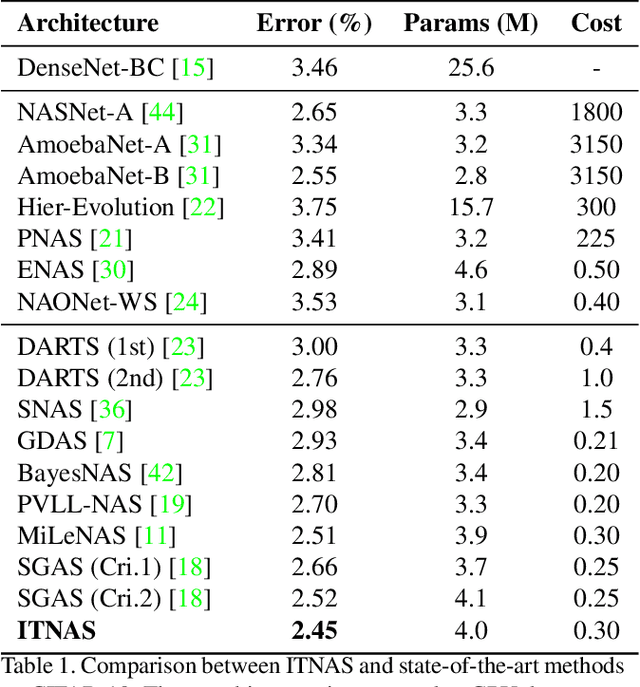
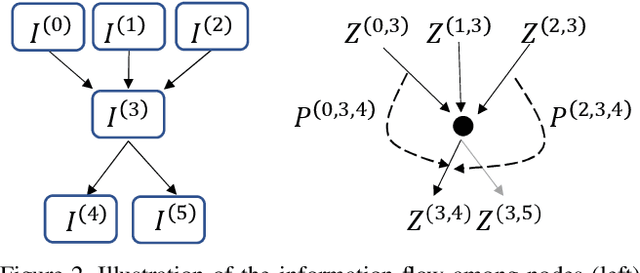
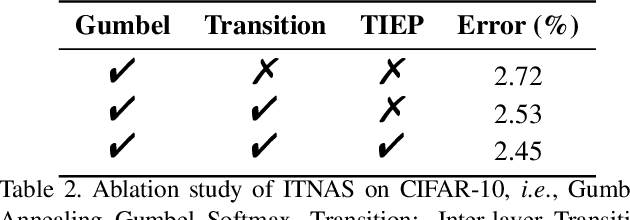
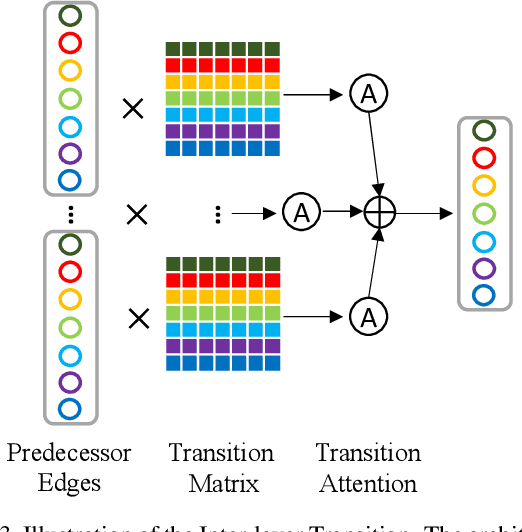
Abstract:Differential Neural Architecture Search (NAS) methods represent the network architecture as a repetitive proxy directed acyclic graph (DAG) and optimize the network weights and architecture weights alternatively in a differential manner. However, existing methods model the architecture weights on each edge (i.e., a layer in the network) as statistically independent variables, ignoring the dependency between edges in DAG induced by their directed topological connections. In this paper, we make the first attempt to investigate such dependency by proposing a novel Inter-layer Transition NAS method. It casts the architecture optimization into a sequential decision process where the dependency between the architecture weights of connected edges is explicitly modeled. Specifically, edges are divided into inner and outer groups according to whether or not their predecessor edges are in the same cell. While the architecture weights of outer edges are optimized independently, those of inner edges are derived sequentially based on the architecture weights of their predecessor edges and the learnable transition matrices in an attentive probability transition manner. Experiments on five benchmarks confirm the value of modeling inter-layer dependency and demonstrate the proposed method outperforms state-of-the-art methods.
Autonomous Deep Learning: A Genetic DCNN Designer for Image Classification
Jul 01, 2018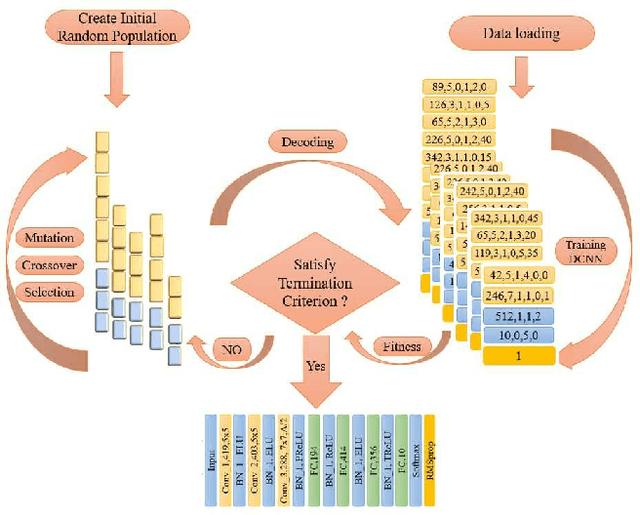
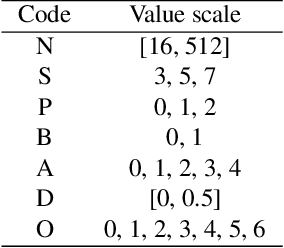
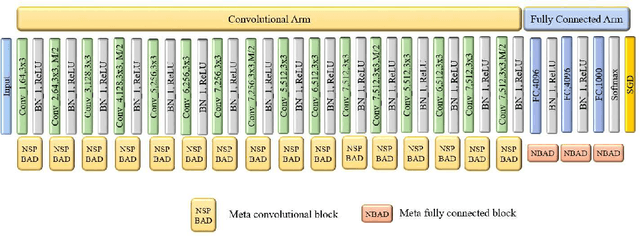
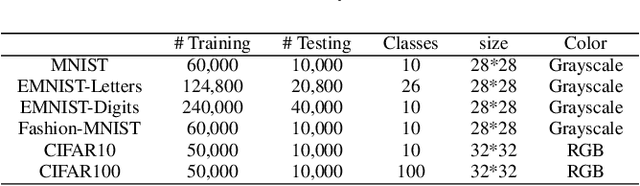
Abstract:Recent years have witnessed the breakthrough success of deep convolutional neural networks (DCNNs) in image classification and other vision applications. Although freeing users from the troublesome handcrafted feature extraction by providing a uniform feature extraction-classification framework, DCNNs still require a handcrafted design of their architectures. In this paper, we propose the genetic DCNN designer, an autonomous learning algorithm can generate a DCNN architecture automatically based on the data available for a specific image classification problem. We first partition a DCNN into multiple stacked meta convolutional blocks and fully connected blocks, each containing the operations of convolution, pooling, fully connection, batch normalization, activation and drop out, and thus convert the architecture into an integer vector. Then, we use refined evolutionary operations, including selection, mutation and crossover to evolve a population of DCNN architectures. Our results on the MNIST, Fashion-MNIST, EMNISTDigit, EMNIST-Letter, CIFAR10 and CIFAR100 datasets suggest that the proposed genetic DCNN designer is able to produce automatically DCNN architectures, whose performance is comparable to, if not better than, that of stateof- the-art DCNN models
A Tribe Competition-Based Genetic Algorithm for Feature Selection in Pattern Classification
Apr 28, 2017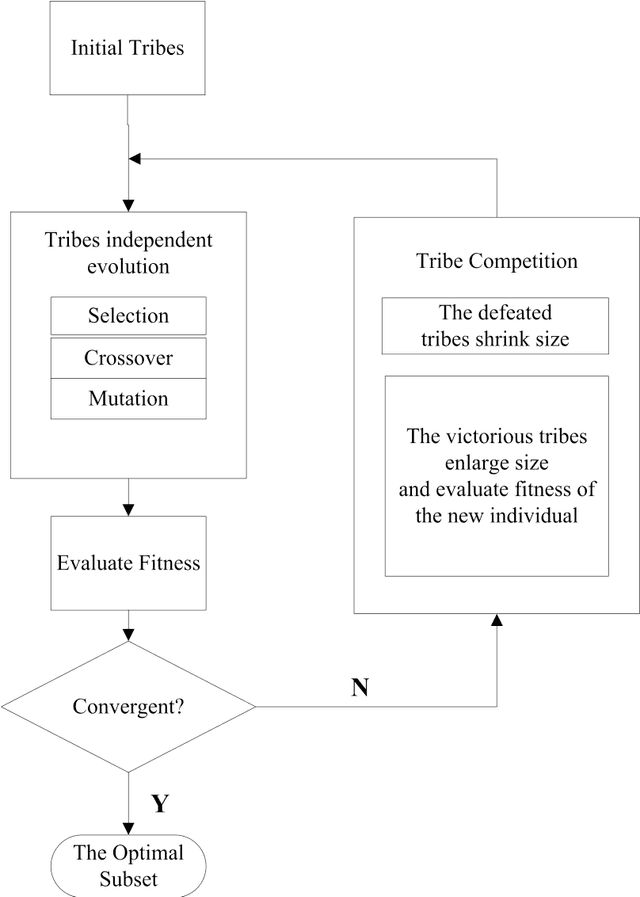
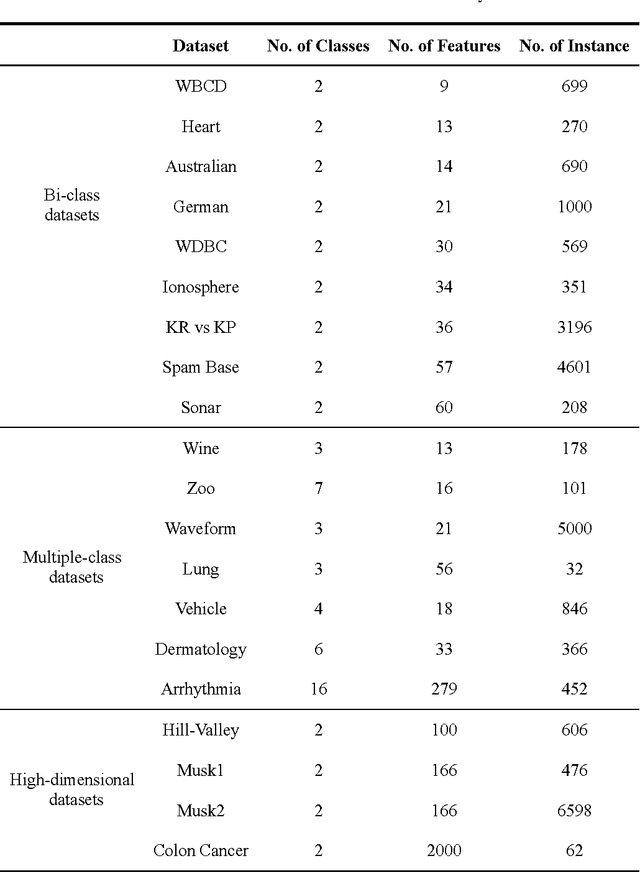
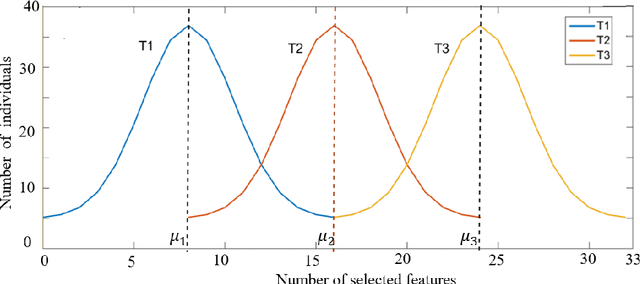
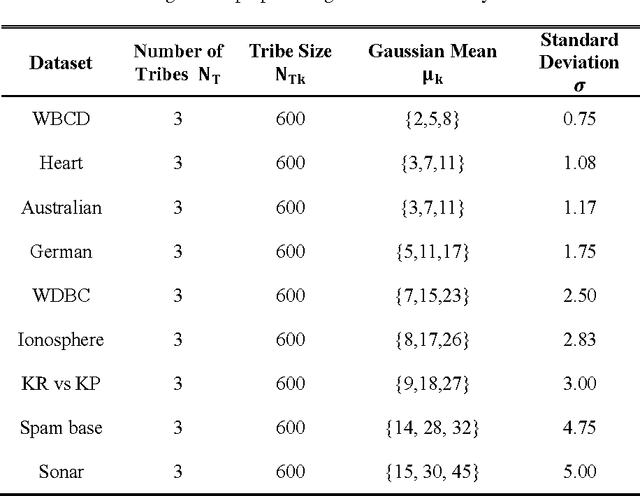
Abstract:Feature selection has always been a critical step in pattern recognition, in which evolutionary algorithms, such as the genetic algorithm (GA), are most commonly used. However, the individual encoding scheme used in various GAs would either pose a bias on the solution or require a pre-specified number of features, and hence may lead to less accurate results. In this paper, a tribe competition-based genetic algorithm (TCbGA) is proposed for feature selection in pattern classification. The population of individuals is divided into multiple tribes, and the initialization and evolutionary operations are modified to ensure that the number of selected features in each tribe follows a Gaussian distribution. Thus each tribe focuses on exploring a specific part of the solution space. Meanwhile, tribe competition is introduced to the evolution process, which allows the winning tribes, which produce better individuals, to enlarge their sizes, i.e. having more individuals to search their parts of the solution space. This algorithm, therefore, avoids the bias on solutions and requirement of a pre-specified number of features. We have evaluated our algorithm against several state-of-the-art feature selection approaches on 20 benchmark datasets. Our results suggest that the proposed TCbGA algorithm can identify the optimal feature subset more effectively and produce more accurate pattern classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge