Asoke K. Nandi
Representation Discrepancy Bridging Method for Remote Sensing Image-Text Retrieval
May 22, 2025Abstract:Remote Sensing Image-Text Retrieval (RSITR) plays a critical role in geographic information interpretation, disaster monitoring, and urban planning by establishing semantic associations between image and textual descriptions. Existing Parameter-Efficient Fine-Tuning (PEFT) methods for Vision-and-Language Pre-training (VLP) models typically adopt symmetric adapter structures for exploring cross-modal correlations. However, the strong discriminative nature of text modality may dominate the optimization process and inhibits image representation learning. The nonnegligible imbalanced cross-modal optimization remains a bottleneck to enhancing the model performance. To address this issue, this study proposes a Representation Discrepancy Bridging (RDB) method for the RSITR task. On the one hand, a Cross-Modal Asymmetric Adapter (CMAA) is designed to enable modality-specific optimization and improve feature alignment. The CMAA comprises a Visual Enhancement Adapter (VEA) and a Text Semantic Adapter (TSA). VEA mines fine-grained image features by Differential Attention (DA) mechanism, while TSA identifies key textual semantics through Hierarchical Attention (HA) mechanism. On the other hand, this study extends the traditional single-task retrieval framework to a dual-task optimization framework and develops a Dual-Task Consistency Loss (DTCL). The DTCL improves cross-modal alignment robustness through an adaptive weighted combination of cross-modal, classification, and exponential moving average consistency constraints. Experiments on RSICD and RSITMD datasets show that the proposed RDB method achieves a 6%-11% improvement in mR metrics compared to state-of-the-art PEFT methods and a 1.15%-2% improvement over the full fine-tuned GeoRSCLIP model.
TEC-Net: Vision Transformer Embrace Convolutional Neural Networks for Medical Image Segmentation
Jun 07, 2023



Abstract:The hybrid architecture of convolution neural networks (CNN) and Transformer has been the most popular method for medical image segmentation. However, the existing networks based on the hybrid architecture suffer from two problems. First, although the CNN branch can capture image local features by using convolution operation, the vanilla convolution is unable to achieve adaptive extraction of image features. Second, although the Transformer branch can model the global information of images, the conventional self-attention only focuses on the spatial self-attention of images and ignores the channel and cross-dimensional self-attention leading to low segmentation accuracy for medical images with complex backgrounds. To solve these problems, we propose vision Transformer embrace convolutional neural networks for medical image segmentation (TEC-Net). Our network has two advantages. First, dynamic deformable convolution (DDConv) is designed in the CNN branch, which not only overcomes the difficulty of adaptive feature extraction using fixed-size convolution kernels, but also solves the defect that different inputs share the same convolution kernel parameters, effectively improving the feature expression ability of CNN branch. Second, in the Transformer branch, a (shifted)-window adaptive complementary attention module ((S)W-ACAM) and compact convolutional projection are designed to enable the network to fully learn the cross-dimensional long-range dependency of medical images with few parameters and calculations. Experimental results show that the proposed TEC-Net provides better medical image segmentation results than SOTA methods including CNN and Transformer networks. In addition, our TEC-Net requires fewer parameters and computational costs and does not rely on pre-training. The code is publicly available at https://github.com/SR0920/TEC-Net.
Lightweight Structure-aware Transformer Network for VHR Remote Sensing Image Change Detection
Jun 03, 2023



Abstract:Popular Transformer networks have been successfully applied to remote sensing (RS) image change detection (CD) identifications and achieve better results than most convolutional neural networks (CNNs), but they still suffer from two main problems. First, the computational complexity of the Transformer grows quadratically with the increase of image spatial resolution, which is unfavorable to very high-resolution (VHR) RS images. Second, these popular Transformer networks tend to ignore the importance of fine-grained features, which results in poor edge integrity and internal tightness for largely changed objects and leads to the loss of small changed objects. To address the above issues, this Letter proposes a Lightweight Structure-aware Transformer (LSAT) network for RS image CD. The proposed LSAT has two advantages. First, a Cross-dimension Interactive Self-attention (CISA) module with linear complexity is designed to replace the vanilla self-attention in visual Transformer, which effectively reduces the computational complexity while improving the feature representation ability of the proposed LSAT. Second, a Structure-aware Enhancement Module (SAEM) is designed to enhance difference features and edge detail information, which can achieve double enhancement by difference refinement and detail aggregation so as to obtain fine-grained features of bi-temporal RS images. Experimental results show that the proposed LSAT achieves significant improvement in detection accuracy and offers a better tradeoff between accuracy and computational costs than most state-of-the-art CD methods for VHR RS images.
Medical Image Segmentation Using Deep Learning: A Survey
Sep 28, 2020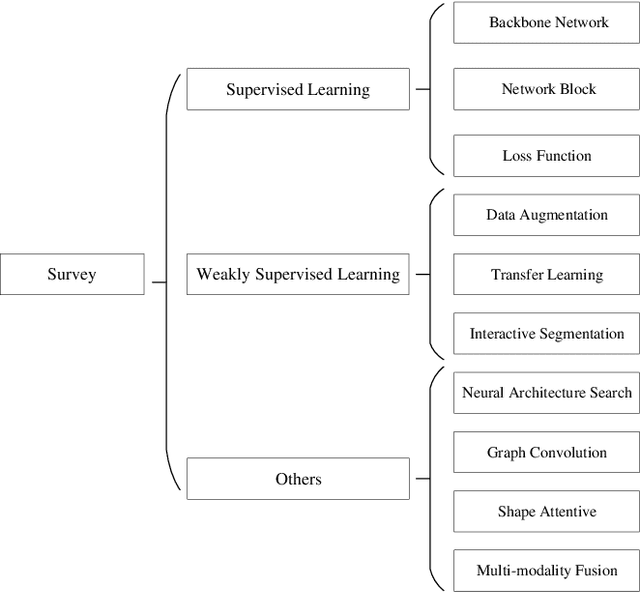
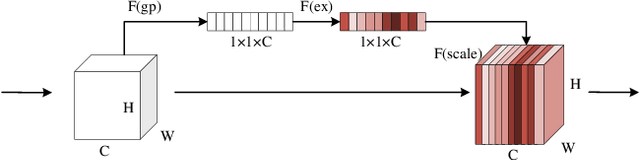
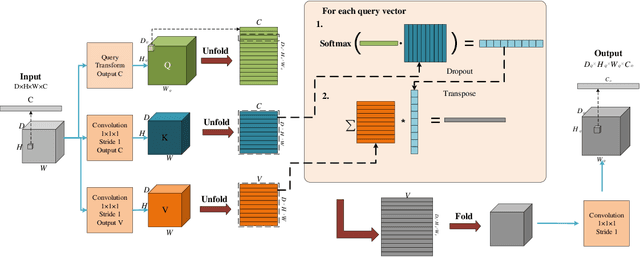
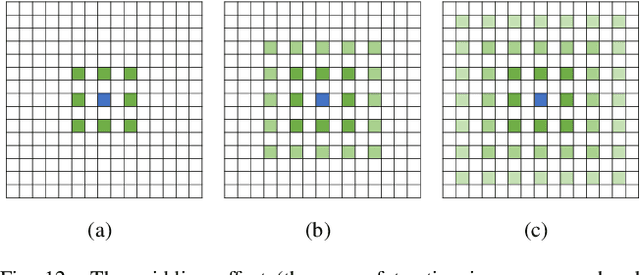
Abstract:Deep learning has been widely used for medical image segmentation and a large number of papers has been presented recording the success of deep learning in the field. In this paper, we present a comprehensive thematic survey on medical image segmentation using deep learning techniques. This paper makes two original contributions. Firstly, compared to traditional surveys that directly divide literatures of deep learning on medical image segmentation into many groups and introduce literatures in detail for each group, we classify currently popular literatures according to a multi-level structure from coarse to fine. Secondly, this paper focuses on supervised and weakly supervised learning approaches, without including unsupervised approaches since they have been introduced in many old surveys and they are not popular currently. For supervised learning approaches, we analyze literatures in three aspects: the selection of backbone networks, the design of network blocks, and the improvement of loss functions. For weakly supervised learning approaches, we investigate literature according to data augmentation, transfer learning, and interactive segmentation, separately. Compared to existing surveys, this survey classifies the literatures very differently from before and is more convenient for readers to understand the relevant rationale and will guide them to think of appropriate improvements in medical image segmentation based on deep learning approaches.
Adaptive Morphological Reconstruction for Seeded Image Segmentation
Apr 08, 2019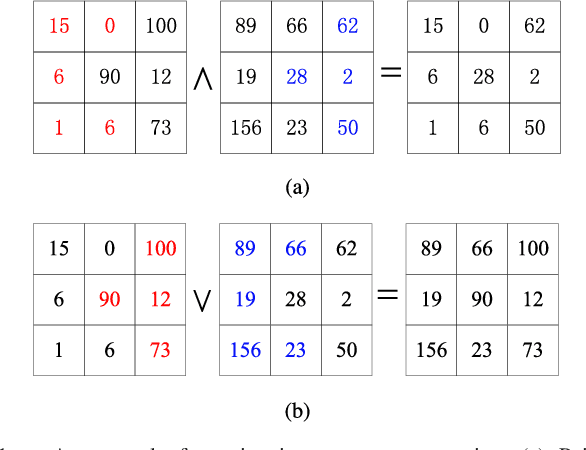
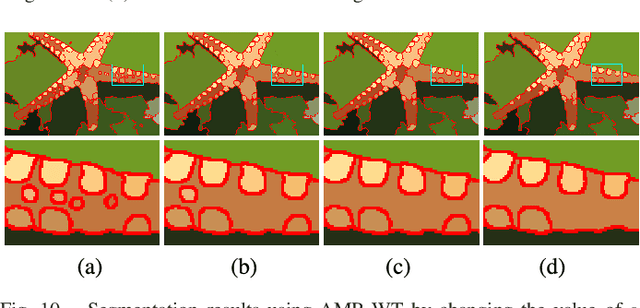
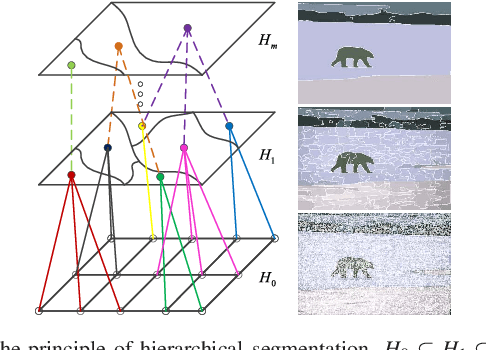

Abstract:Morphological reconstruction (MR) is often employed by seeded image segmentation algorithms such as watershed transform and power watershed as it is able to filter seeds (regional minima) to reduce over-segmentation. However, MR might mistakenly filter meaningful seeds that are required for generating accurate segmentation and it is also sensitive to the scale because a single-scale structuring element is employed. In this paper, a novel adaptive morphological reconstruction (AMR) operation is proposed that has three advantages. Firstly, AMR can adaptively filter useless seeds while preserving meaningful ones. Secondly, AMR is insensitive to the scale of structuring elements because multiscale structuring elements are employed. Finally, AMR has two attractive properties: monotonic increasingness and convergence that help seeded segmentation algorithms to achieve a hierarchical segmentation. Experiments clearly demonstrate that AMR is useful for improving algorithms of seeded image segmentation and seed-based spectral segmentation. Compared to several state-of-the-art algorithms, the proposed algorithms provide better segmentation results requiring less computing time. Source code is available at https://github.com/SUST-reynole/AMR.
Diffusion map for clustering fMRI spatial maps extracted by independent component analysis
Sep 27, 2013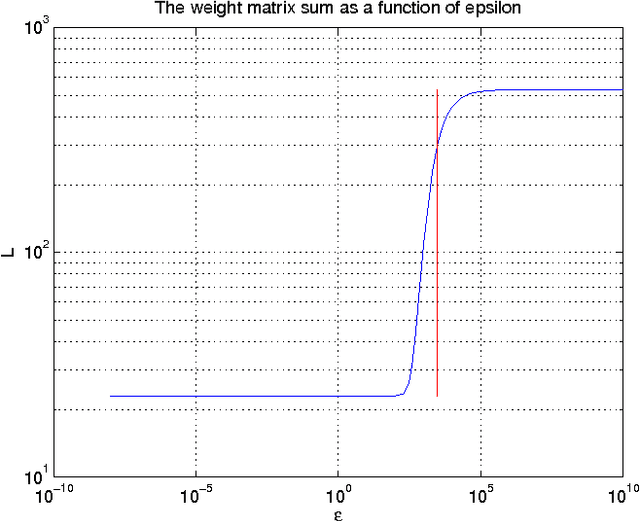

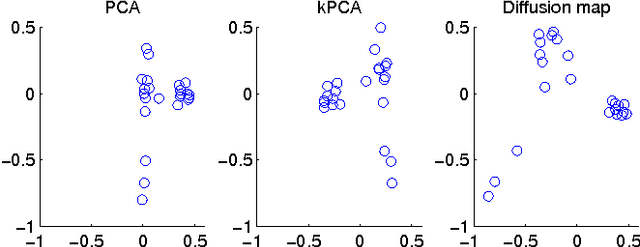

Abstract:Functional magnetic resonance imaging (fMRI) produces data about activity inside the brain, from which spatial maps can be extracted by independent component analysis (ICA). In datasets, there are n spatial maps that contain p voxels. The number of voxels is very high compared to the number of analyzed spatial maps. Clustering of the spatial maps is usually based on correlation matrices. This usually works well, although such a similarity matrix inherently can explain only a certain amount of the total variance contained in the high-dimensional data where n is relatively small but p is large. For high-dimensional space, it is reasonable to perform dimensionality reduction before clustering. In this research, we used the recently developed diffusion map for dimensionality reduction in conjunction with spectral clustering. This research revealed that the diffusion map based clustering worked as well as the more traditional methods, and produced more compact clusters when needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge