Fengyu Cong
Key-Value Pair-Free Continual Learner via Task-Specific Prompt-Prototype
Jan 08, 2026Abstract:Continual learning aims to enable models to acquire new knowledge while retaining previously learned information. Prompt-based methods have shown remarkable performance in this domain; however, they typically rely on key-value pairing, which can introduce inter-task interference and hinder scalability. To overcome these limitations, we propose a novel approach employing task-specific Prompt-Prototype (ProP), thereby eliminating the need for key-value pairs. In our method, task-specific prompts facilitate more effective feature learning for the current task, while corresponding prototypes capture the representative features of the input. During inference, predictions are generated by binding each task-specific prompt with its associated prototype. Additionally, we introduce regularization constraints during prompt initialization to penalize excessively large values, thereby enhancing stability. Experiments on several widely used datasets demonstrate the effectiveness of the proposed method. In contrast to mainstream prompt-based approaches, our framework removes the dependency on key-value pairs, offering a fresh perspective for future continual learning research.
Multi-modal Knowledge Decomposition based Online Distillation for Biomarker Prediction in Breast Cancer Histopathology
Aug 24, 2025Abstract:Immunohistochemical (IHC) biomarker prediction benefits from multi-modal data fusion analysis. However, the simultaneous acquisition of multi-modal data, such as genomic and pathological information, is often challenging due to cost or technical limitations. To address this challenge, we propose an online distillation approach based on Multi-modal Knowledge Decomposition (MKD) to enhance IHC biomarker prediction in haematoxylin and eosin (H\&E) stained histopathology images. This method leverages paired genomic-pathology data during training while enabling inference using either pathology slides alone or both modalities. Two teacher and one student models are developed to extract modality-specific and modality-general features by minimizing the MKD loss. To maintain the internal structural relationships between samples, Similarity-preserving Knowledge Distillation (SKD) is applied. Additionally, Collaborative Learning for Online Distillation (CLOD) facilitates mutual learning between teacher and student models, encouraging diverse and complementary learning dynamics. Experiments on the TCGA-BRCA and in-house QHSU datasets demonstrate that our approach achieves superior performance in IHC biomarker prediction using uni-modal data. Our code is available at https://github.com/qiyuanzz/MICCAI2025_MKD.
FCNCP: A Coupled Nonnegative CANDECOMP/PARAFAC Decomposition Based on Federated Learning
Apr 18, 2024



Abstract:In the field of brain science, data sharing across servers is becoming increasingly challenging due to issues such as industry competition, privacy security, and administrative procedure policies and regulations. Therefore, there is an urgent need to develop new methods for data analysis and processing that enable scientific collaboration without data sharing. In view of this, this study proposes to study and develop a series of efficient non-negative coupled tensor decomposition algorithm frameworks based on federated learning called FCNCP for the EEG data arranged on different servers. It combining the good discriminative performance of tensor decomposition in high-dimensional data representation and decomposition, the advantages of coupled tensor decomposition in cross-sample tensor data analysis, and the features of federated learning for joint modelling in distributed servers. The algorithm utilises federation learning to establish coupling constraints for data distributed across different servers. In the experiments, firstly, simulation experiments are carried out using simulated data, and stable and consistent decomposition results are obtained, which verify the effectiveness of the proposed algorithms in this study. Then the FCNCP algorithm was utilised to decompose the fifth-order event-related potential (ERP) tensor data collected by applying proprioceptive stimuli on the left and right hands. It was found that contralateral stimulation induced more symmetrical components in the activation areas of the left and right hemispheres. The conclusions drawn are consistent with the interpretations of related studies in cognitive neuroscience, demonstrating that the method can efficiently process higher-order EEG data and that some key hidden information can be preserved.
Fast Learnings of Coupled Nonnegative Tensor Decomposition Using Optimal Gradient and Low-rank Approximation
Feb 10, 2023Abstract:Nonnegative tensor decomposition has been widely applied in signal processing and neuroscience, etc. When it comes to group analysis of multi-block tensors, traditional tensor decomposition is insufficient to utilize the shared/similar information among tensors. In this study, we propose a coupled nonnegative CANDECOMP/PARAFAC decomposition algorithm optimized by the alternating proximal gradient method (CoNCPDAPG), which is capable of a simultaneous decomposition of tensors from different samples that are partially linked and a simultaneous extraction of common components, individual components and core tensors. Due to the low optimization efficiency brought by the nonnegative constraint and the high-dimensional nature of the data, we further propose the lraCoNCPD-APG algorithm by combining low-rank approximation and the proposed CoNCPD-APG method. When processing multi-block large-scale tensors, the proposed lraCoNCPD-APG algorithm can greatly reduce the computational load without compromising the decomposition quality. Experiment results of coupled nonnegative tensor decomposition problems designed for synthetic data, real-world face images and event-related potential data demonstrate the practicability and superiority of the proposed algorithms.
Sparse Nonnegative CANDECOMP/PARAFAC Decomposition in Block Coordinate Descent Framework: A Comparison Study
Dec 27, 2018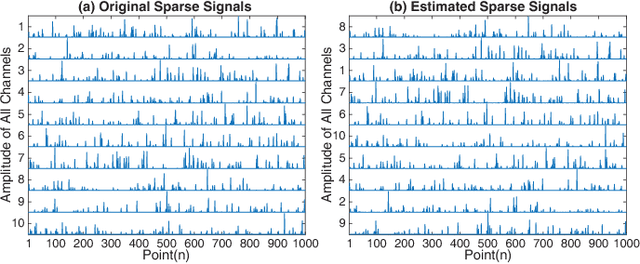
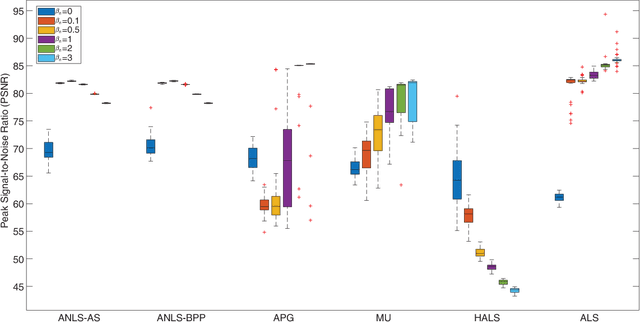
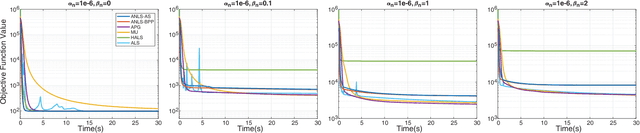
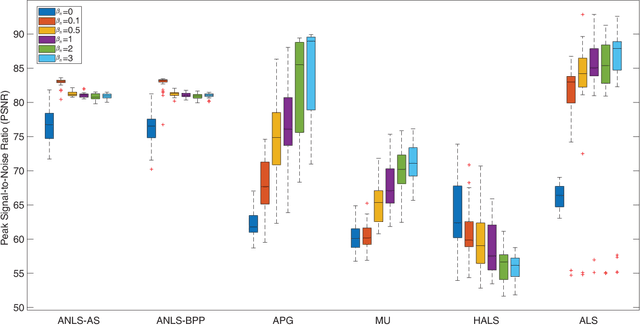
Abstract:Nonnegative CANDECOMP/PARAFAC (NCP) decomposition is an important tool to process nonnegative tensor. Sometimes, additional sparse regularization is needed to extract meaningful nonnegative and sparse components. Thus, an optimization method for NCP that can impose sparsity efficiently is required. In this paper, we construct NCP with sparse regularization (sparse NCP) by l1-norm. Several popular optimization methods in block coordinate descent framework are employed to solve the sparse NCP, all of which are deeply analyzed with mathematical solutions. We compare these methods by experiments on synthetic and real tensor data, both of which contain third-order and fourth-order cases. After comparison, the methods that have fast computation and high effectiveness to impose sparsity will be concluded. In addition, we proposed an accelerated method to compute the objective function and relative error of sparse NCP, which has significantly improved the computation of tensor decomposition especially for higher-order tensor.
Diffusion map for clustering fMRI spatial maps extracted by independent component analysis
Sep 27, 2013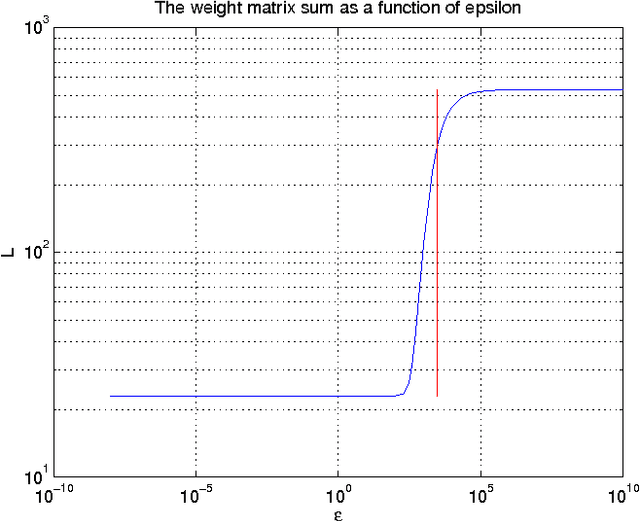

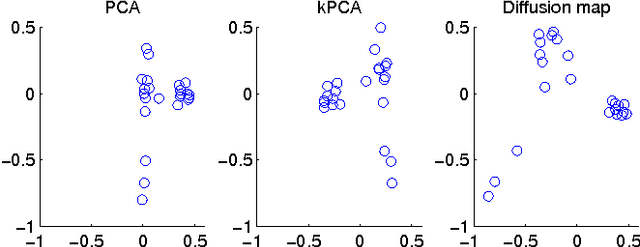

Abstract:Functional magnetic resonance imaging (fMRI) produces data about activity inside the brain, from which spatial maps can be extracted by independent component analysis (ICA). In datasets, there are n spatial maps that contain p voxels. The number of voxels is very high compared to the number of analyzed spatial maps. Clustering of the spatial maps is usually based on correlation matrices. This usually works well, although such a similarity matrix inherently can explain only a certain amount of the total variance contained in the high-dimensional data where n is relatively small but p is large. For high-dimensional space, it is reasonable to perform dimensionality reduction before clustering. In this research, we used the recently developed diffusion map for dimensionality reduction in conjunction with spectral clustering. This research revealed that the diffusion map based clustering worked as well as the more traditional methods, and produced more compact clusters when needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge