Arzucan Özgür
Department of Computer Engineering Boğaziçi University
evoBPE: Evolutionary Protein Sequence Tokenization
Mar 11, 2025Abstract:Recent advancements in computational biology have drawn compelling parallels between protein sequences and linguistic structures, highlighting the need for sophisticated tokenization methods that capture the intricate evolutionary dynamics of protein sequences. Current subword tokenization techniques, primarily developed for natural language processing, often fail to represent protein sequences' complex structural and functional properties adequately. This study introduces evoBPE, a novel tokenization approach that integrates evolutionary mutation patterns into sequence segmentation, addressing critical limitations in existing methods. By leveraging established substitution matrices, evoBPE transcends traditional frequency-based tokenization strategies. The method generates candidate token pairs through biologically informed mutations, evaluating them based on pairwise alignment scores and frequency thresholds. Extensive experiments on human protein sequences show that evoBPE performs better across multiple dimensions. Domain conservation analysis reveals that evoBPE consistently outperforms standard Byte-Pair Encoding, particularly as vocabulary size increases. Furthermore, embedding similarity analysis using ESM-2 suggests that mutation-based token replacements preserve biological sequence properties more effectively than arbitrary substitutions. The research contributes to protein sequence representation by introducing a mutation-aware tokenization method that better captures evolutionary nuances. By bridging computational linguistics and molecular biology, evoBPE opens new possibilities for machine learning applications in protein function prediction, structural modeling, and evolutionary analysis.
Linguistic Laws Meet Protein Sequences: A Comparative Analysis of Subword Tokenization Methods
Nov 26, 2024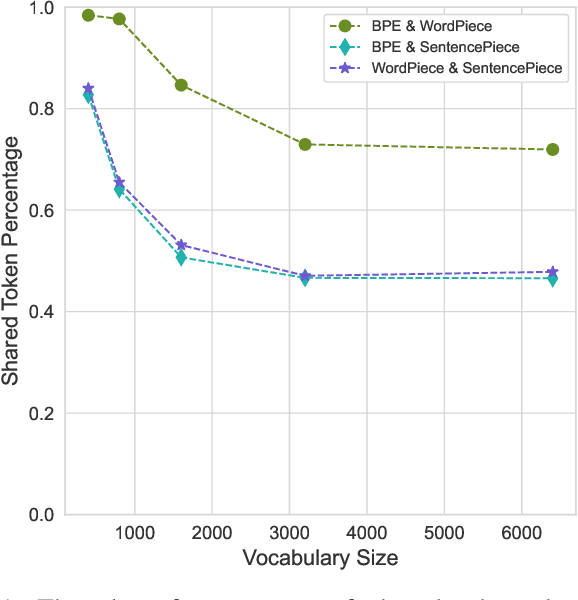
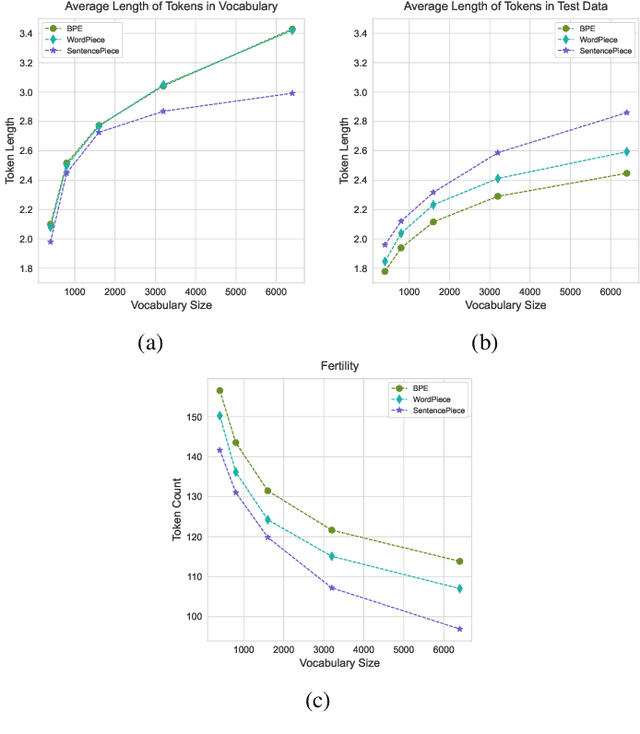
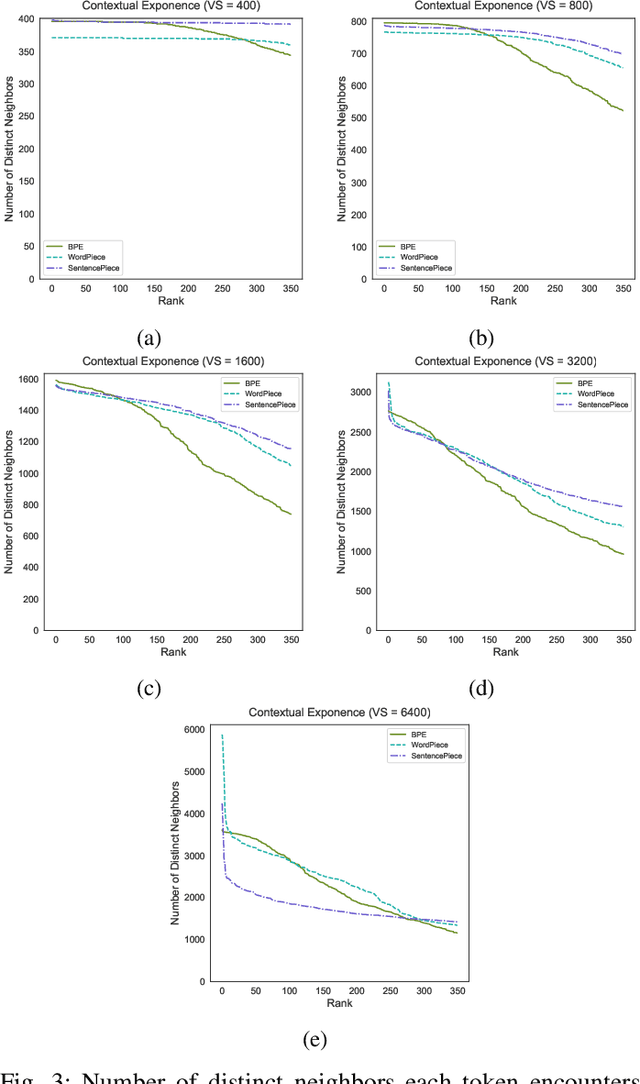
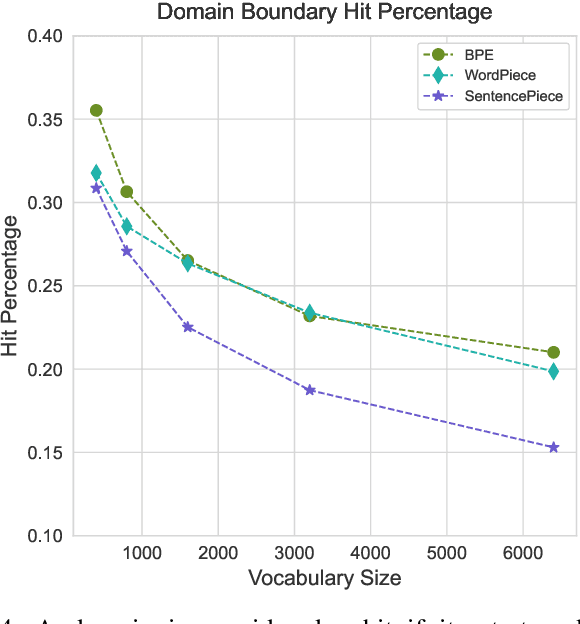
Abstract:Tokenization is a crucial step in processing protein sequences for machine learning models, as proteins are complex sequences of amino acids that require meaningful segmentation to capture their functional and structural properties. However, existing subword tokenization methods, developed primarily for human language, may be inadequate for protein sequences, which have unique patterns and constraints. This study evaluates three prominent tokenization approaches, Byte-Pair Encoding (BPE), WordPiece, and SentencePiece, across varying vocabulary sizes (400-6400), analyzing their effectiveness in protein sequence representation, domain boundary preservation, and adherence to established linguistic laws. Our comprehensive analysis reveals distinct behavioral patterns among these tokenizers, with vocabulary size significantly influencing their performance. BPE demonstrates better contextual specialization and marginally better domain boundary preservation at smaller vocabularies, while SentencePiece achieves better encoding efficiency, leading to lower fertility scores. WordPiece offers a balanced compromise between these characteristics. However, all tokenizers show limitations in maintaining protein domain integrity, particularly as vocabulary size increases. Analysis of linguistic law adherence shows partial compliance with Zipf's and Brevity laws but notable deviations from Menzerath's law, suggesting that protein sequences may follow distinct organizational principles from natural languages. These findings highlight the limitations of applying traditional NLP tokenization methods to protein sequences and emphasize the need for developing specialized tokenization strategies that better account for the unique characteristics of proteins.
Evaluation of GPT and BERT-based models on identifying protein-protein interactions in biomedical text
Mar 30, 2023Abstract:Detecting protein-protein interactions (PPIs) is crucial for understanding genetic mechanisms, disease pathogenesis, and drug design. However, with the fast-paced growth of biomedical literature, there is a growing need for automated and accurate extraction of PPIs to facilitate scientific knowledge discovery. Pre-trained language models, such as generative pre-trained transformer (GPT) and bidirectional encoder representations from transformers (BERT), have shown promising results in natural language processing (NLP) tasks. We evaluated the PPI identification performance of various GPT and BERT models using a manually curated benchmark corpus of 164 PPIs in 77 sentences from learning language in logic (LLL). BERT-based models achieved the best overall performance, with PubMedBERT achieving the highest precision (85.17%) and F1-score (86.47%) and BioM-ALBERT achieving the highest recall (93.83%). Despite not being explicitly trained for biomedical texts, GPT-4 achieved comparable performance to the best BERT models with 83.34% precision, 76.57% recall, and 79.18% F1-score. These findings suggest that GPT models can effectively detect PPIs from text data and have the potential for use in biomedical literature mining tasks.
Explain to Me: Towards Understanding Privacy Decisions
Jan 05, 2023Abstract:Privacy assistants help users manage their privacy online. Their tasks could vary from detecting privacy violations to recommending sharing actions for content that the user intends to share. Recent work on these tasks are promising and show that privacy assistants can successfully tackle them. However, for such privacy assistants to be employed by users, it is important that these assistants can explain their decisions to users. Accordingly, this paper develops a methodology to create explanations of privacy. The methodology is based on identifying important topics in a domain of interest, providing explanation schemes for decisions, and generating them automatically. We apply our proposed methodology on a real-world privacy data set, which contains images labeled as private or public to explain the labels. We evaluate our approach on a user study that depicts what factors are influential for users to find explanations useful.
Exploiting Pretrained Biochemical Language Models for Targeted Drug Design
Sep 02, 2022
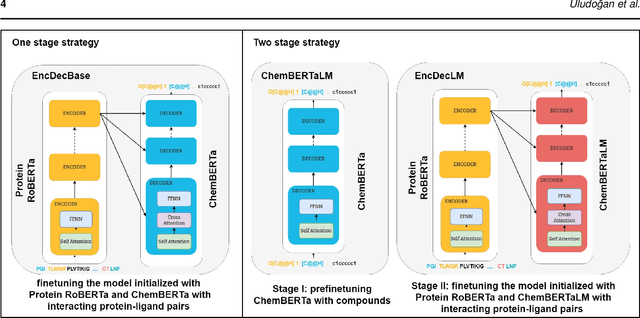
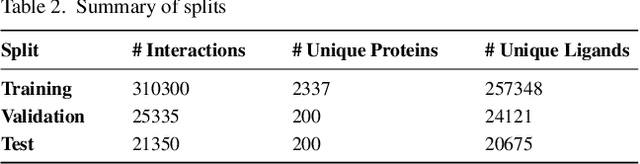
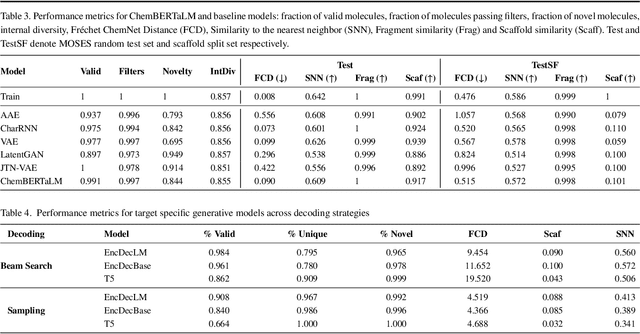
Abstract:Motivation: The development of novel compounds targeting proteins of interest is one of the most important tasks in the pharmaceutical industry. Deep generative models have been applied to targeted molecular design and have shown promising results. Recently, target-specific molecule generation has been viewed as a translation between the protein language and the chemical language. However, such a model is limited by the availability of interacting protein-ligand pairs. On the other hand, large amounts of unlabeled protein sequences and chemical compounds are available and have been used to train language models that learn useful representations. In this study, we propose exploiting pretrained biochemical language models to initialize (i.e. warm start) targeted molecule generation models. We investigate two warm start strategies: (i) a one-stage strategy where the initialized model is trained on targeted molecule generation (ii) a two-stage strategy containing a pre-finetuning on molecular generation followed by target specific training. We also compare two decoding strategies to generate compounds: beam search and sampling. Results: The results show that the warm-started models perform better than a baseline model trained from scratch. The two proposed warm-start strategies achieve similar results to each other with respect to widely used metrics from benchmarks. However, docking evaluation of the generated compounds for a number of novel proteins suggests that the one-stage strategy generalizes better than the two-stage strategy. Additionally, we observe that beam search outperforms sampling in both docking evaluation and benchmark metrics for assessing compound quality. Availability and implementation: The source code is available at https://github.com/boun-tabi/biochemical-lms-for-drug-design and the materials are archived in Zenodo at https://doi.org/10.5281/zenodo.6832145
Enhancements to the BOUN Treebank Reflecting the Agglutinative Nature of Turkish
Jul 24, 2022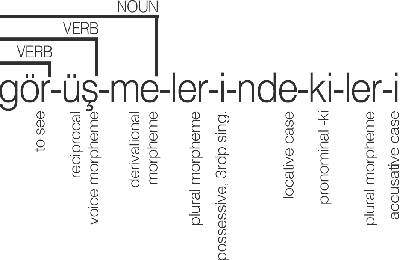
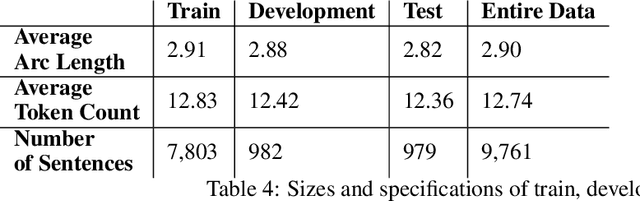
Abstract:In this study, we aim to offer linguistically motivated solutions to resolve the issues of the lack of representation of null morphemes, highly productive derivational processes, and syncretic morphemes of Turkish in the BOUN Treebank without diverging from the Universal Dependencies framework. In order to tackle these issues, new annotation conventions were introduced by splitting certain lemmas and employing the MISC (miscellaneous) tab in the UD framework to denote derivation. Representational capabilities of the re-annotated treebank were tested on a LSTM-based dependency parser and an updated version of the BoAT Tool is introduced.
A Self-aware Personal Assistant for Making Personalized Privacy Decisions
May 18, 2022


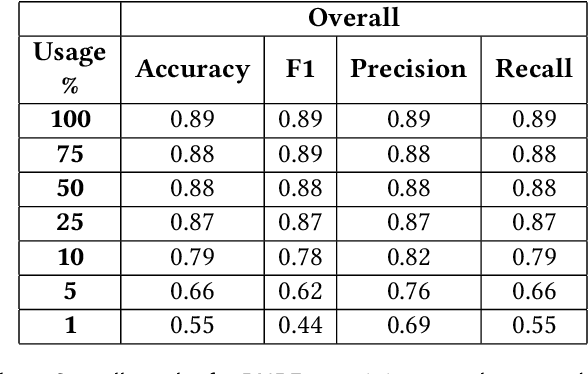
Abstract:Many software systems, such as online social networks enable users to share information about themselves. While the action of sharing is simple, it requires an elaborate thought process on privacy: what to share, with whom to share, and for what purposes. Thinking about these for each piece of content to be shared is tedious. Recent approaches to tackle this problem build personal assistants that can help users by learning what is private over time and recommending privacy labels such as private or public to individual content that a user considers sharing. However, privacy is inherently ambiguous and highly personal. Existing approaches to recommend privacy decisions do not address these aspects of privacy sufficiently. Ideally, a personal assistant should be able to adjust its recommendation based on a given user, considering that user's privacy understanding. Moreover, the personal assistant should be able to assess when its recommendation would be uncertain and let the user make the decision on her own. Accordingly, this paper proposes a personal assistant that uses evidential deep learning to classify content based on its privacy label. An important characteristic of the personal assistant is that it can model its uncertainty in its decisions explicitly, determine that it does not know the answer, and delegate from making a recommendation when its uncertainty is high. By factoring in the user's own understanding of privacy, such as risk factors or own labels, the personal assistant can personalize its recommendations per user. We evaluate our proposed personal assistant using a well-known data set. Our results show that our personal assistant can accurately identify uncertain cases, personalize them to its user's needs, and thus helps users preserve their privacy well.
A Dataset and BERT-based Models for Targeted Sentiment Analysis on Turkish Texts
May 09, 2022
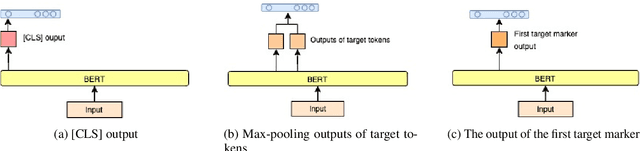


Abstract:Targeted Sentiment Analysis aims to extract sentiment towards a particular target from a given text. It is a field that is attracting attention due to the increasing accessibility of the Internet, which leads people to generate an enormous amount of data. Sentiment analysis, which in general requires annotated data for training, is a well-researched area for widely studied languages such as English. For low-resource languages such as Turkish, there is a lack of such annotated data. We present an annotated Turkish dataset suitable for targeted sentiment analysis. We also propose BERT-based models with different architectures to accomplish the task of targeted sentiment analysis. The results demonstrate that the proposed models outperform the traditional sentiment analysis models for the targeted sentiment analysis task.
Cluster-based Mention Typing for Named Entity Disambiguation
Sep 23, 2021

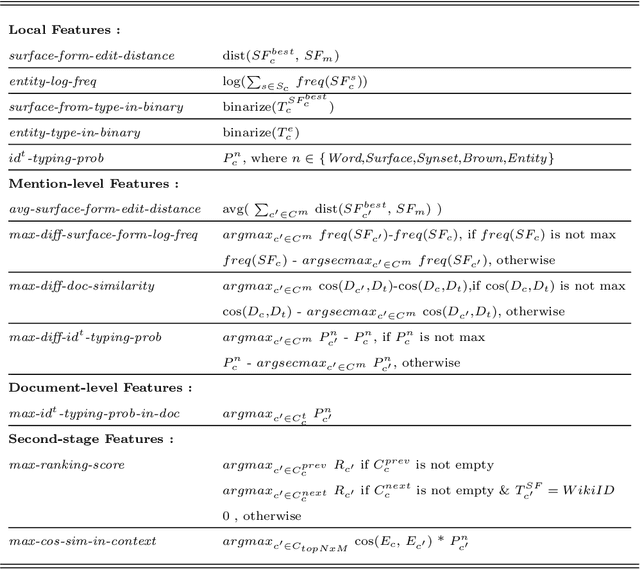
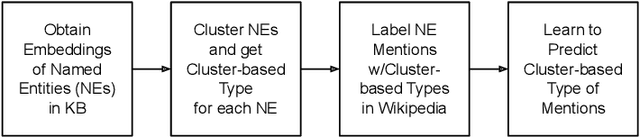
Abstract:An entity mention in text such as "Washington" may correspond to many different named entities such as the city "Washington D.C." or the newspaper "Washington Post." The goal of named entity disambiguation is to identify the mentioned named entity correctly among all possible candidates. If the type (e.g. location or person) of a mentioned entity can be correctly predicted from the context, it may increase the chance of selecting the right candidate by assigning low probability to the unlikely ones. This paper proposes cluster-based mention typing for named entity disambiguation. The aim of mention typing is to predict the type of a given mention based on its context. Generally, manually curated type taxonomies such as Wikipedia categories are used. We introduce cluster-based mention typing, where named entities are clustered based on their contextual similarities and the cluster ids are assigned as types. The hyperlinked mentions and their context in Wikipedia are used in order to obtain these cluster-based types. Then, mention typing models are trained on these mentions, which have been labeled with their cluster-based types through distant supervision. At the named entity disambiguation phase, first the cluster-based types of a given mention are predicted and then, these types are used as features in a ranking model to select the best entity among the candidates. We represent entities at multiple contextual levels and obtain different clusterings (and thus typing models) based on each level. As each clustering breaks the entity space differently, mention typing based on each clustering discriminates the mention differently. When predictions from all typing models are used together, our system achieves better or comparable results based on randomization tests with respect to the state-of-the-art levels on four defacto test sets.
* 46 pages, 11 figures, 14 tables
Balancing Methods for Multi-label Text Classification with Long-Tailed Class Distribution
Sep 10, 2021

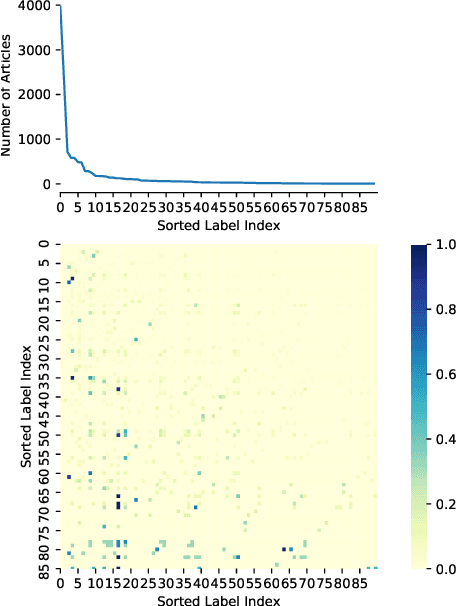
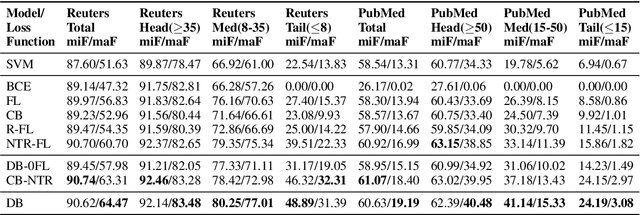
Abstract:Multi-label text classification is a challenging task because it requires capturing label dependencies. It becomes even more challenging when class distribution is long-tailed. Resampling and re-weighting are common approaches used for addressing the class imbalance problem, however, they are not effective when there is label dependency besides class imbalance because they result in oversampling of common labels. Here, we introduce the application of balancing loss functions for multi-label text classification. We perform experiments on a general domain dataset with 90 labels (Reuters-21578) and a domain-specific dataset from PubMed with 18211 labels. We find that a distribution-balanced loss function, which inherently addresses both the class imbalance and label linkage problems, outperforms commonly used loss functions. Distribution balancing methods have been successfully used in the image recognition field. Here, we show their effectiveness in natural language processing. Source code is available at https://github.com/blessu/BalancedLossNLP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge