Gökçe Uludoğan
TURNA: A Turkish Encoder-Decoder Language Model for Enhanced Understanding and Generation
Jan 25, 2024Abstract:The recent advances in natural language processing have predominantly favored well-resourced English-centric models, resulting in a significant gap with low-resource languages. In this work, we introduce the language model TURNA, which is developed for the low-resource language Turkish and is capable of both natural language understanding and generation tasks. TURNA is pretrained with an encoder-decoder architecture based on the unified framework UL2 with a diverse corpus that we specifically curated for this purpose. We evaluated TURNA with three generation tasks and five understanding tasks for Turkish. The results show that TURNA outperforms several multilingual models in both understanding and generation tasks, and competes with monolingual Turkish models in understanding tasks. TURNA is made available at https://huggingface.co/boun-tabi-LMG/TURNA .
Exploiting Pretrained Biochemical Language Models for Targeted Drug Design
Sep 02, 2022
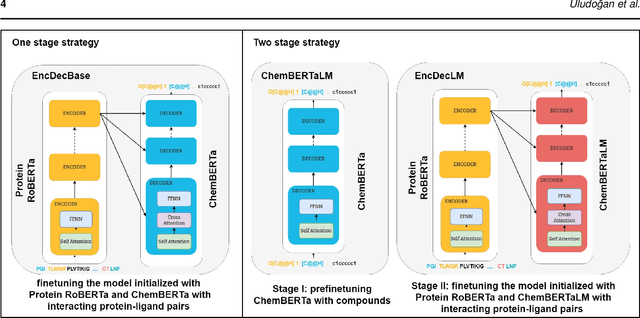
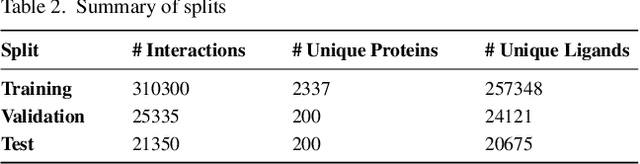
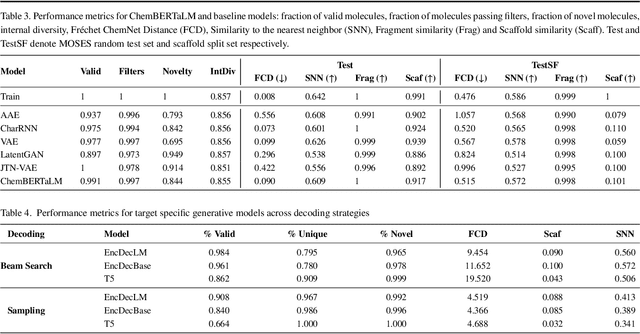
Abstract:Motivation: The development of novel compounds targeting proteins of interest is one of the most important tasks in the pharmaceutical industry. Deep generative models have been applied to targeted molecular design and have shown promising results. Recently, target-specific molecule generation has been viewed as a translation between the protein language and the chemical language. However, such a model is limited by the availability of interacting protein-ligand pairs. On the other hand, large amounts of unlabeled protein sequences and chemical compounds are available and have been used to train language models that learn useful representations. In this study, we propose exploiting pretrained biochemical language models to initialize (i.e. warm start) targeted molecule generation models. We investigate two warm start strategies: (i) a one-stage strategy where the initialized model is trained on targeted molecule generation (ii) a two-stage strategy containing a pre-finetuning on molecular generation followed by target specific training. We also compare two decoding strategies to generate compounds: beam search and sampling. Results: The results show that the warm-started models perform better than a baseline model trained from scratch. The two proposed warm-start strategies achieve similar results to each other with respect to widely used metrics from benchmarks. However, docking evaluation of the generated compounds for a number of novel proteins suggests that the one-stage strategy generalizes better than the two-stage strategy. Additionally, we observe that beam search outperforms sampling in both docking evaluation and benchmark metrics for assessing compound quality. Availability and implementation: The source code is available at https://github.com/boun-tabi/biochemical-lms-for-drug-design and the materials are archived in Zenodo at https://doi.org/10.5281/zenodo.6832145
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge