Burak Suyunu
evoBPE: Evolutionary Protein Sequence Tokenization
Mar 11, 2025Abstract:Recent advancements in computational biology have drawn compelling parallels between protein sequences and linguistic structures, highlighting the need for sophisticated tokenization methods that capture the intricate evolutionary dynamics of protein sequences. Current subword tokenization techniques, primarily developed for natural language processing, often fail to represent protein sequences' complex structural and functional properties adequately. This study introduces evoBPE, a novel tokenization approach that integrates evolutionary mutation patterns into sequence segmentation, addressing critical limitations in existing methods. By leveraging established substitution matrices, evoBPE transcends traditional frequency-based tokenization strategies. The method generates candidate token pairs through biologically informed mutations, evaluating them based on pairwise alignment scores and frequency thresholds. Extensive experiments on human protein sequences show that evoBPE performs better across multiple dimensions. Domain conservation analysis reveals that evoBPE consistently outperforms standard Byte-Pair Encoding, particularly as vocabulary size increases. Furthermore, embedding similarity analysis using ESM-2 suggests that mutation-based token replacements preserve biological sequence properties more effectively than arbitrary substitutions. The research contributes to protein sequence representation by introducing a mutation-aware tokenization method that better captures evolutionary nuances. By bridging computational linguistics and molecular biology, evoBPE opens new possibilities for machine learning applications in protein function prediction, structural modeling, and evolutionary analysis.
Linguistic Laws Meet Protein Sequences: A Comparative Analysis of Subword Tokenization Methods
Nov 26, 2024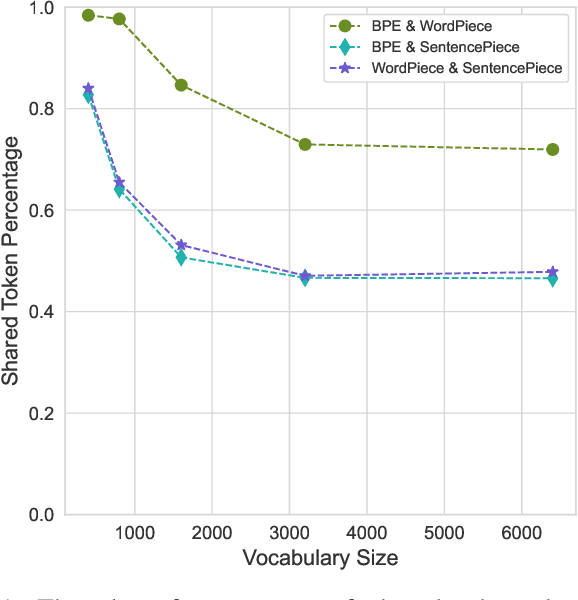
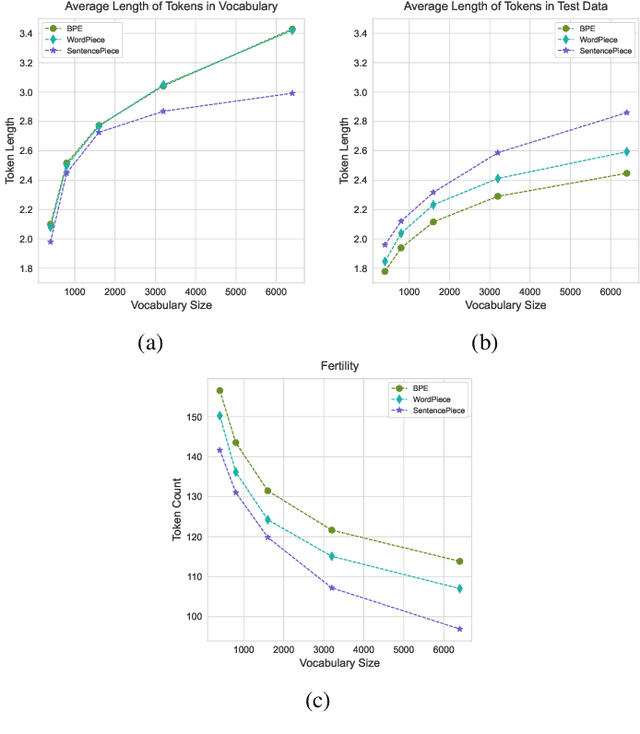
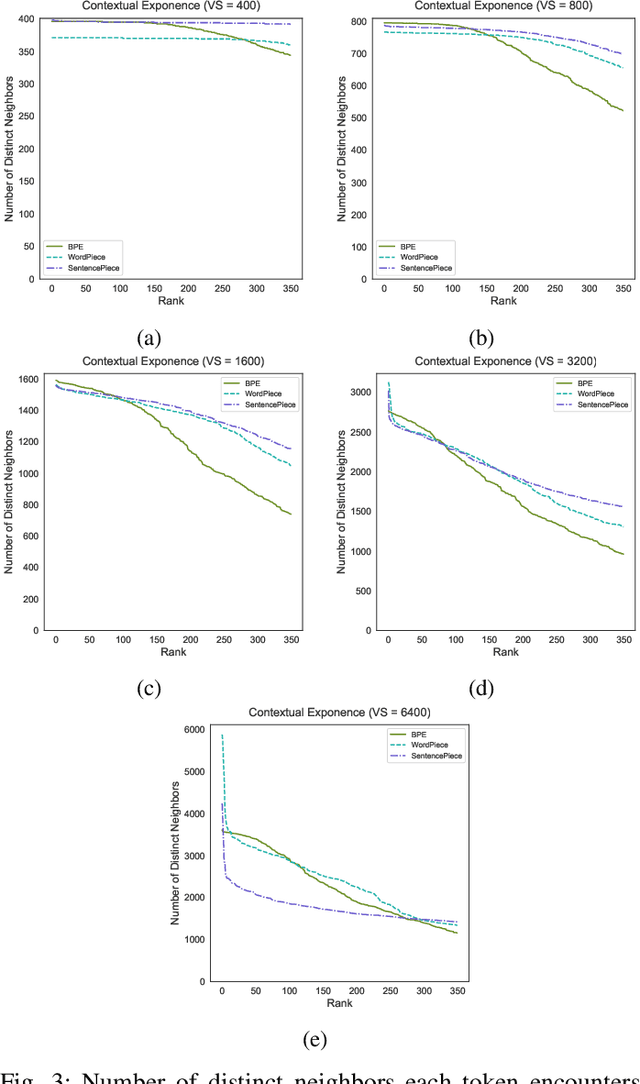
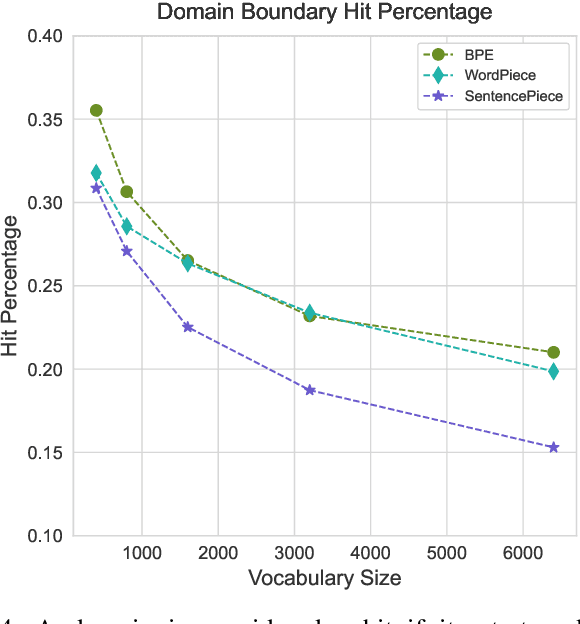
Abstract:Tokenization is a crucial step in processing protein sequences for machine learning models, as proteins are complex sequences of amino acids that require meaningful segmentation to capture their functional and structural properties. However, existing subword tokenization methods, developed primarily for human language, may be inadequate for protein sequences, which have unique patterns and constraints. This study evaluates three prominent tokenization approaches, Byte-Pair Encoding (BPE), WordPiece, and SentencePiece, across varying vocabulary sizes (400-6400), analyzing their effectiveness in protein sequence representation, domain boundary preservation, and adherence to established linguistic laws. Our comprehensive analysis reveals distinct behavioral patterns among these tokenizers, with vocabulary size significantly influencing their performance. BPE demonstrates better contextual specialization and marginally better domain boundary preservation at smaller vocabularies, while SentencePiece achieves better encoding efficiency, leading to lower fertility scores. WordPiece offers a balanced compromise between these characteristics. However, all tokenizers show limitations in maintaining protein domain integrity, particularly as vocabulary size increases. Analysis of linguistic law adherence shows partial compliance with Zipf's and Brevity laws but notable deviations from Menzerath's law, suggesting that protein sequences may follow distinct organizational principles from natural languages. These findings highlight the limitations of applying traditional NLP tokenization methods to protein sequences and emphasize the need for developing specialized tokenization strategies that better account for the unique characteristics of proteins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge