Antoine Simoulin
Enhancing Open-Vocabulary Object Detection through Multi-Level Fine-Grained Visual-Language Alignment
Jan 31, 2026Abstract:Traditional object detection systems are typically constrained to predefined categories, limiting their applicability in dynamic environments. In contrast, open-vocabulary object detection (OVD) enables the identification of objects from novel classes not present in the training set. Recent advances in visual-language modeling have led to significant progress of OVD. However, prior works face challenges in either adapting the single-scale image backbone from CLIP to the detection framework or ensuring robust visual-language alignment. We propose Visual-Language Detection (VLDet), a novel framework that revamps feature pyramid for fine-grained visual-language alignment, leading to improved OVD performance. With the VL-PUB module, VLDet effectively exploits the visual-language knowledge from CLIP and adapts the backbone for object detection through feature pyramid. In addition, we introduce the SigRPN block, which incorporates a sigmoid-based anchor-text contrastive alignment loss to improve detection of novel categories. Through extensive experiments, our approach achieves 58.7 AP for novel classes on COCO2017 and 24.8 AP on LVIS, surpassing all state-of-the-art methods and achieving significant improvements of 27.6% and 6.9%, respectively. Furthermore, VLDet also demonstrates superior zero-shot performance on closed-set object detection.
DocAgent: A Multi-Agent System for Automated Code Documentation Generation
Apr 11, 2025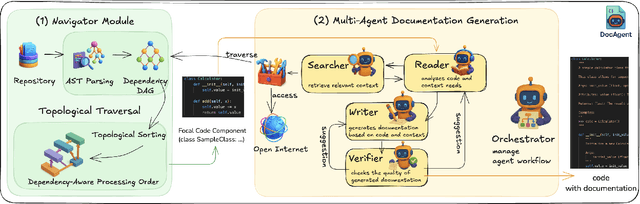
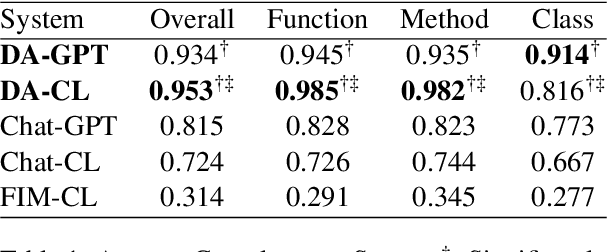
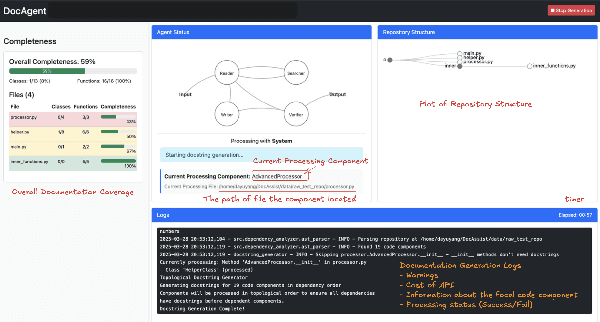

Abstract:High-quality code documentation is crucial for software development especially in the era of AI. However, generating it automatically using Large Language Models (LLMs) remains challenging, as existing approaches often produce incomplete, unhelpful, or factually incorrect outputs. We introduce DocAgent, a novel multi-agent collaborative system using topological code processing for incremental context building. Specialized agents (Reader, Searcher, Writer, Verifier, Orchestrator) then collaboratively generate documentation. We also propose a multi-faceted evaluation framework assessing Completeness, Helpfulness, and Truthfulness. Comprehensive experiments show DocAgent significantly outperforms baselines consistently. Our ablation study confirms the vital role of the topological processing order. DocAgent offers a robust approach for reliable code documentation generation in complex and proprietary repositories.
Code to Think, Think to Code: A Survey on Code-Enhanced Reasoning and Reasoning-Driven Code Intelligence in LLMs
Feb 26, 2025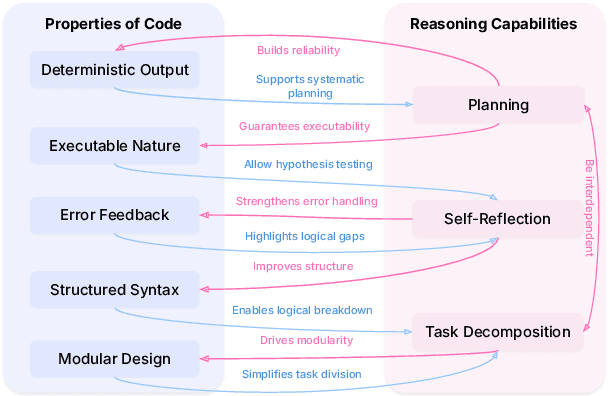
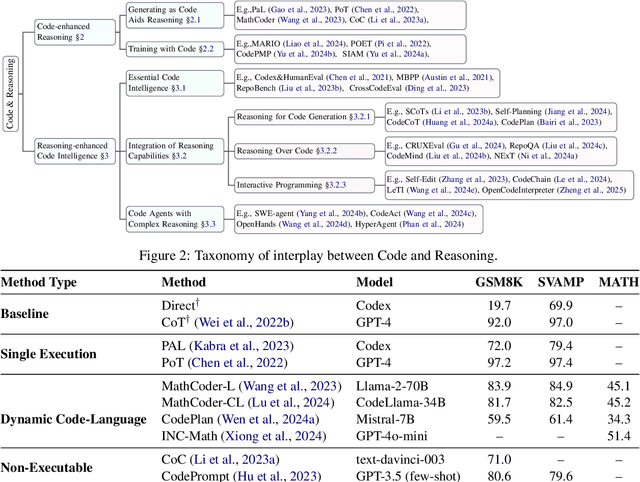


Abstract:In large language models (LLMs), code and reasoning reinforce each other: code offers an abstract, modular, and logic-driven structure that supports reasoning, while reasoning translates high-level goals into smaller, executable steps that drive more advanced code intelligence. In this study, we examine how code serves as a structured medium for enhancing reasoning: it provides verifiable execution paths, enforces logical decomposition, and enables runtime validation. We also explore how improvements in reasoning have transformed code intelligence from basic completion to advanced capabilities, enabling models to address complex software engineering tasks through planning and debugging. Finally, we identify key challenges and propose future research directions to strengthen this synergy, ultimately improving LLM's performance in both areas.
Memory-Efficient Fine-Tuning of Transformers via Token Selection
Jan 31, 2025



Abstract:Fine-tuning provides an effective means to specialize pre-trained models for various downstream tasks. However, fine-tuning often incurs high memory overhead, especially for large transformer-based models, such as LLMs. While existing methods may reduce certain parts of the memory required for fine-tuning, they still require caching all intermediate activations computed in the forward pass to update weights during the backward pass. In this work, we develop TokenTune, a method to reduce memory usage, specifically the memory to store intermediate activations, in the fine-tuning of transformer-based models. During the backward pass, TokenTune approximates the gradient computation by backpropagating through just a subset of input tokens. Thus, with TokenTune, only a subset of intermediate activations are cached during the forward pass. Also, TokenTune can be easily combined with existing methods like LoRA, further reducing the memory cost. We evaluate our approach on pre-trained transformer models with up to billions of parameters, considering the performance on multiple downstream tasks such as text classification and question answering in a few-shot learning setup. Overall, TokenTune achieves performance on par with full fine-tuning or representative memory-efficient fine-tuning methods, while greatly reducing the memory footprint, especially when combined with other methods with complementary memory reduction mechanisms. We hope that our approach will facilitate the fine-tuning of large transformers, in specializing them for specific domains or co-training them with other neural components from a larger system. Our code is available at https://github.com/facebookresearch/tokentune.
GLEMOS: Benchmark for Instantaneous Graph Learning Model Selection
Apr 02, 2024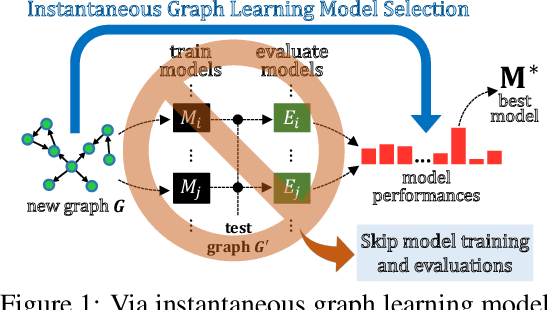
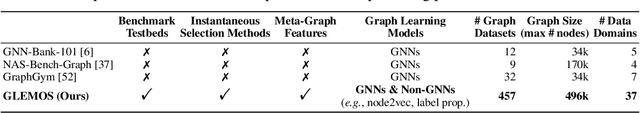
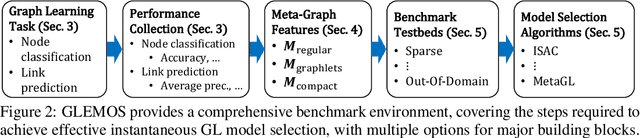
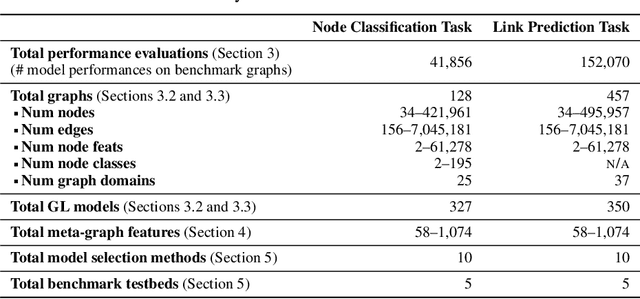
Abstract:The choice of a graph learning (GL) model (i.e., a GL algorithm and its hyperparameter settings) has a significant impact on the performance of downstream tasks. However, selecting the right GL model becomes increasingly difficult and time consuming as more and more GL models are developed. Accordingly, it is of great significance and practical value to equip users of GL with the ability to perform a near-instantaneous selection of an effective GL model without manual intervention. Despite the recent attempts to tackle this important problem, there has been no comprehensive benchmark environment to evaluate the performance of GL model selection methods. To bridge this gap, we present GLEMOS in this work, a comprehensive benchmark for instantaneous GL model selection that makes the following contributions. (i) GLEMOS provides extensive benchmark data for fundamental GL tasks, i.e., link prediction and node classification, including the performances of 366 models on 457 graphs on these tasks. (ii) GLEMOS designs multiple evaluation settings, and assesses how effectively representative model selection techniques perform in these different settings. (iii) GLEMOS is designed to be easily extended with new models, new graphs, and new performance records. (iv) Based on the experimental results, we discuss the limitations of existing approaches and highlight future research directions. To promote research on this significant problem, we make the benchmark data and code publicly available at https://github.com/facebookresearch/glemos.
Forward Learning of Graph Neural Networks
Mar 16, 2024Abstract:Graph neural networks (GNNs) have achieved remarkable success across a wide range of applications, such as recommendation, drug discovery, and question answering. Behind the success of GNNs lies the backpropagation (BP) algorithm, which is the de facto standard for training deep neural networks (NNs). However, despite its effectiveness, BP imposes several constraints, which are not only biologically implausible, but also limit the scalability, parallelism, and flexibility in learning NNs. Examples of such constraints include storage of neural activities computed in the forward pass for use in the subsequent backward pass, and the dependence of parameter updates on non-local signals. To address these limitations, the forward-forward algorithm (FF) was recently proposed as an alternative to BP in the image classification domain, which trains NNs by performing two forward passes over positive and negative data. Inspired by this advance, we propose ForwardGNN in this work, a new forward learning procedure for GNNs, which avoids the constraints imposed by BP via an effective layer-wise local forward training. ForwardGNN extends the original FF to deal with graph data and GNNs, and makes it possible to operate without generating negative inputs (hence no longer forward-forward). Further, ForwardGNN enables each layer to learn from both the bottom-up and top-down signals without relying on the backpropagation of errors. Extensive experiments on real-world datasets show the effectiveness and generality of the proposed forward graph learning framework. We release our code at https://github.com/facebookresearch/forwardgnn.
An innovative solution for breast cancer textual big data analysis
Dec 06, 2017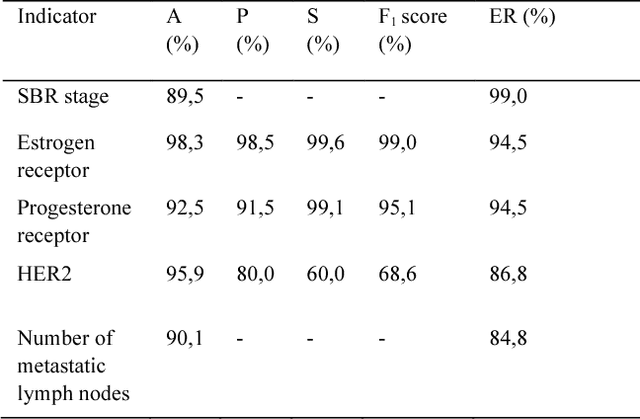
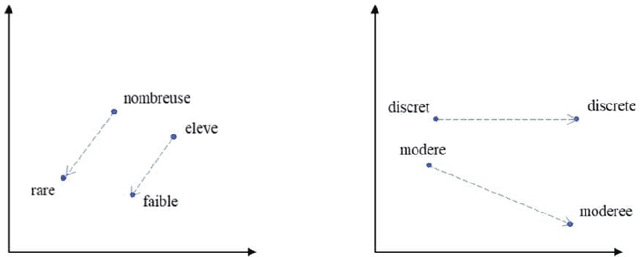
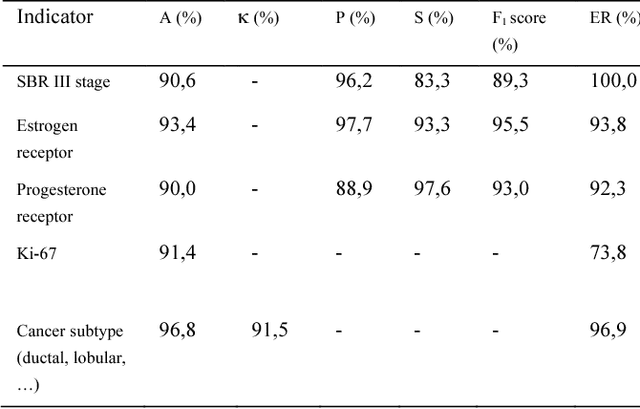
Abstract:The digitalization of stored information in hospitals now allows for the exploitation of medical data in text format, as electronic health records (EHRs), initially gathered for other purposes than epidemiology. Manual search and analysis operations on such data become tedious. In recent years, the use of natural language processing (NLP) tools was highlighted to automatize the extraction of information contained in EHRs, structure it and perform statistical analysis on this structured information. The main difficulties with the existing approaches is the requirement of synonyms or ontology dictionaries, that are mostly available in English only and do not include local or custom notations. In this work, a team composed of oncologists as domain experts and data scientists develop a custom NLP-based system to process and structure textual clinical reports of patients suffering from breast cancer. The tool relies on the combination of standard text mining techniques and an advanced synonym detection method. It allows for a global analysis by retrieval of indicators such as medical history, tumor characteristics, therapeutic responses, recurrences and prognosis. The versatility of the method allows to obtain easily new indicators, thus opening up the way for retrospective studies with a substantial reduction of the amount of manual work. With no need for biomedical annotators or pre-defined ontologies, this language-agnostic method reached an good extraction accuracy for several concepts of interest, according to a comparison with a manually structured file, without requiring any existing corpus with local or new notations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge