Nicolas Bousquet
EDF R\&D PRISME, SINCLAIR AI Lab, LPSM
WTNN: Weibull-Tailored Neural Networks for survival analysis
Dec 09, 2025Abstract:The Weibull distribution is a commonly adopted choice for modeling the survival of systems subject to maintenance over time. When only proxy indicators and censored observations are available, it becomes necessary to express the distribution's parameters as functions of time-dependent covariates. Deep neural networks provide the flexibility needed to learn complex relationships between these covariates and operational lifetime, thereby extending the capabilities of traditional regression-based models. Motivated by the analysis of a fleet of military vehicles operating in highly variable and demanding environments, as well as by the limitations observed in existing methodologies, this paper introduces WTNN, a new neural network-based modeling framework specifically designed for Weibull survival studies. The proposed architecture is specifically designed to incorporate qualitative prior knowledge regarding the most influential covariates, in a manner consistent with the shape and structure of the Weibull distribution. Through numerical experiments, we show that this approach can be reliably trained on proxy and right-censored data, and is capable of producing robust and interpretable survival predictions that can improve existing approaches.
Explaining Models under Multivariate Bernoulli Distribution via Hoeffding Decomposition
Oct 08, 2025Abstract:Explaining the behavior of predictive models with random inputs can be achieved through sub-models decomposition, where such sub-models have easier interpretable features. Arising from the uncertainty quantification community, recent results have demonstrated the existence and uniqueness of a generalized Hoeffding decomposition for such predictive models when the stochastic input variables are correlated, based on concepts of oblique projection onto L 2 subspaces. This article focuses on the case where the input variables have Bernoulli distributions and provides a complete description of this decomposition. We show that in this case the underlying L 2 subspaces are one-dimensional and that the functional decomposition is explicit. This leads to a complete interpretability framework and theoretically allows reverse engineering. Explicit indicators of the influence of inputs on the output prediction (exemplified by Sobol' indices and Shapley effects) can be explicitly derived. Illustrated by numerical experiments, this type of analysis proves useful for addressing decision-support problems, based on binary decision diagrams, Boolean networks or binary neural networks. The article outlines perspectives for exploring high-dimensional settings and, beyond the case of binary inputs, extending these findings to models with finite countable inputs.
Benchmarking XAI Explanations with Human-Aligned Evaluations
Nov 04, 2024
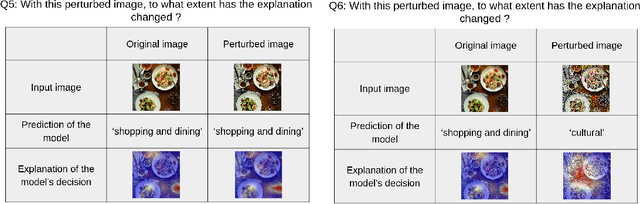

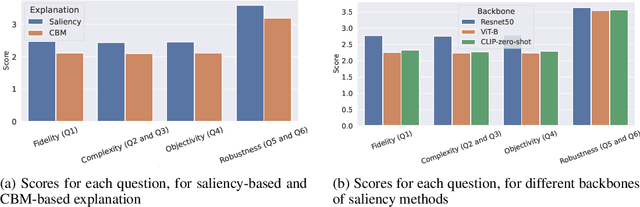
Abstract:In this paper, we introduce PASTA (Perceptual Assessment System for explanaTion of Artificial intelligence), a novel framework for a human-centric evaluation of XAI techniques in computer vision. Our first key contribution is a human evaluation of XAI explanations on four diverse datasets (COCO, Pascal Parts, Cats Dogs Cars, and MonumAI) which constitutes the first large-scale benchmark dataset for XAI, with annotations at both the image and concept levels. This dataset allows for robust evaluation and comparison across various XAI methods. Our second major contribution is a data-based metric for assessing the interpretability of explanations. It mimics human preferences, based on a database of human evaluations of explanations in the PASTA-dataset. With its dataset and metric, the PASTA framework provides consistent and reliable comparisons between XAI techniques, in a way that is scalable but still aligned with human evaluations. Additionally, our benchmark allows for comparisons between explanations across different modalities, an aspect previously unaddressed. Our findings indicate that humans tend to prefer saliency maps over other explanation types. Moreover, we provide evidence that human assessments show a low correlation with existing XAI metrics that are numerically simulated by probing the model.
Understanding black-box models with dependent inputs through a generalization of Hoeffding's decomposition
Oct 10, 2023Abstract:One of the main challenges for interpreting black-box models is the ability to uniquely decompose square-integrable functions of non-mutually independent random inputs into a sum of functions of every possible subset of variables. However, dealing with dependencies among inputs can be complicated. We propose a novel framework to study this problem, linking three domains of mathematics: probability theory, functional analysis, and combinatorics. We show that, under two reasonable assumptions on the inputs (non-perfect functional dependence and non-degenerate stochastic dependence), it is always possible to decompose uniquely such a function. This ``canonical decomposition'' is relatively intuitive and unveils the linear nature of non-linear functions of non-linearly dependent inputs. In this framework, we effectively generalize the well-known Hoeffding decomposition, which can be seen as a particular case. Oblique projections of the black-box model allow for novel interpretability indices for evaluation and variance decomposition. Aside from their intuitive nature, the properties of these novel indices are studied and discussed. This result offers a path towards a more precise uncertainty quantification, which can benefit sensitivity analyses and interpretability studies, whenever the inputs are dependent. This decomposition is illustrated analytically, and the challenges to adopting these results in practice are discussed.
Quantile-constrained Wasserstein projections for robust interpretability of numerical and machine learning models
Sep 23, 2022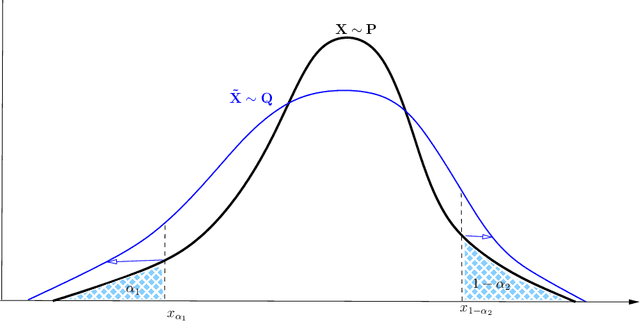
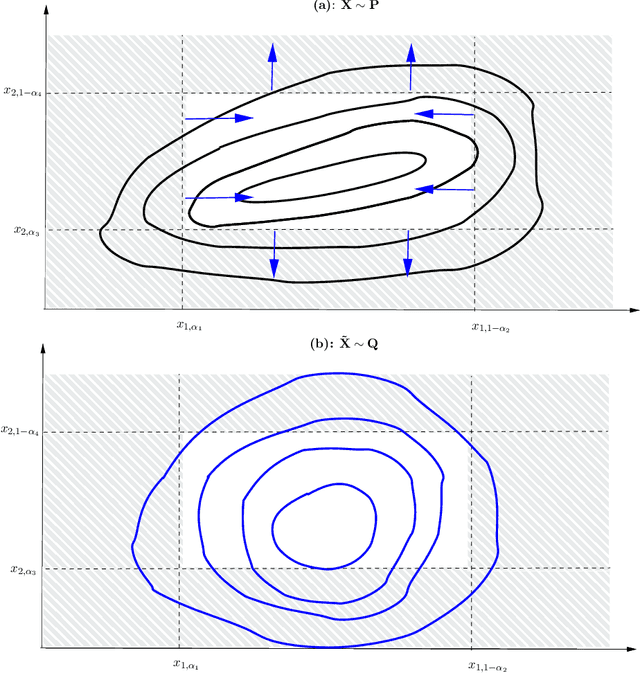
Abstract:Robustness studies of black-box models is recognized as a necessary task for numerical models based on structural equations and predictive models learned from data. These studies must assess the model's robustness to possible misspecification of regarding its inputs (e.g., covariate shift). The study of black-box models, through the prism of uncertainty quantification (UQ), is often based on sensitivity analysis involving a probabilistic structure imposed on the inputs, while ML models are solely constructed from observed data. Our work aim at unifying the UQ and ML interpretability approaches, by providing relevant and easy-to-use tools for both paradigms. To provide a generic and understandable framework for robustness studies, we define perturbations of input information relying on quantile constraints and projections with respect to the Wasserstein distance between probability measures, while preserving their dependence structure. We show that this perturbation problem can be analytically solved. Ensuring regularity constraints by means of isotonic polynomial approximations leads to smoother perturbations, which can be more suitable in practice. Numerical experiments on real case studies, from the UQ and ML fields, highlight the computational feasibility of such studies and provide local and global insights on the robustness of black-box models to input perturbations.
An innovative solution for breast cancer textual big data analysis
Dec 06, 2017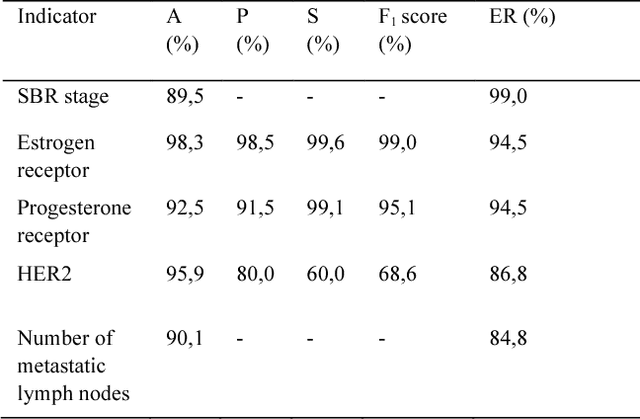
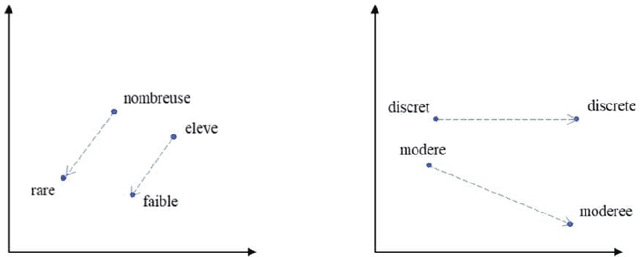
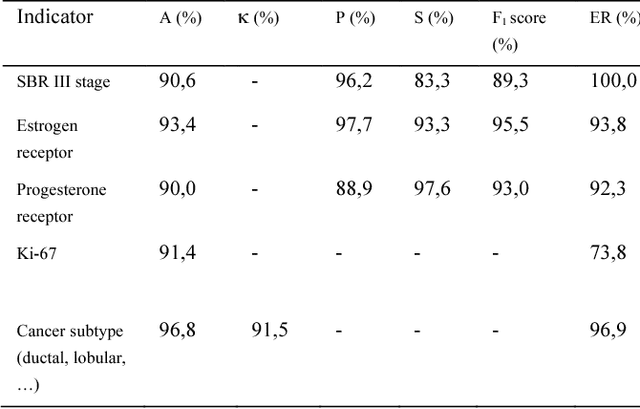
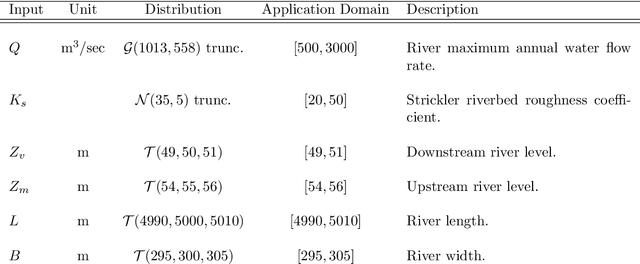
Abstract:The digitalization of stored information in hospitals now allows for the exploitation of medical data in text format, as electronic health records (EHRs), initially gathered for other purposes than epidemiology. Manual search and analysis operations on such data become tedious. In recent years, the use of natural language processing (NLP) tools was highlighted to automatize the extraction of information contained in EHRs, structure it and perform statistical analysis on this structured information. The main difficulties with the existing approaches is the requirement of synonyms or ontology dictionaries, that are mostly available in English only and do not include local or custom notations. In this work, a team composed of oncologists as domain experts and data scientists develop a custom NLP-based system to process and structure textual clinical reports of patients suffering from breast cancer. The tool relies on the combination of standard text mining techniques and an advanced synonym detection method. It allows for a global analysis by retrieval of indicators such as medical history, tumor characteristics, therapeutic responses, recurrences and prognosis. The versatility of the method allows to obtain easily new indicators, thus opening up the way for retrospective studies with a substantial reduction of the amount of manual work. With no need for biomedical annotators or pre-defined ontologies, this language-agnostic method reached an good extraction accuracy for several concepts of interest, according to a comparison with a manually structured file, without requiring any existing corpus with local or new notations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge