An innovative solution for breast cancer textual big data analysis
Paper and Code
Dec 06, 2017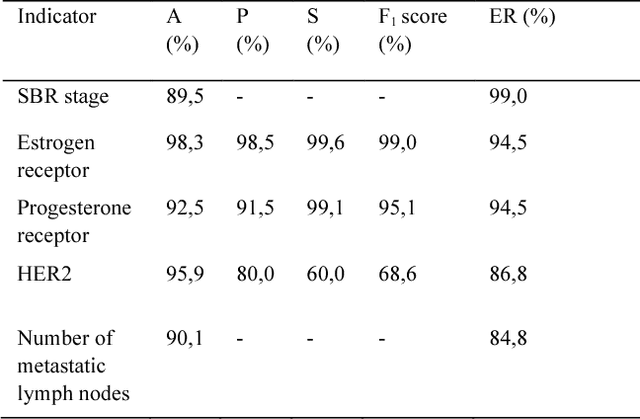
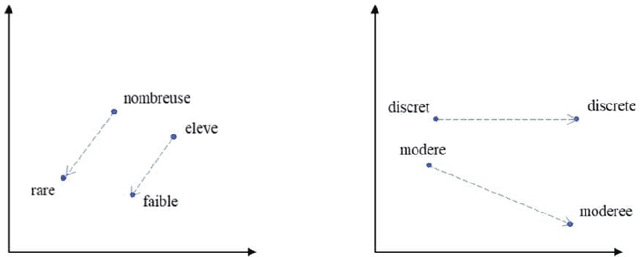
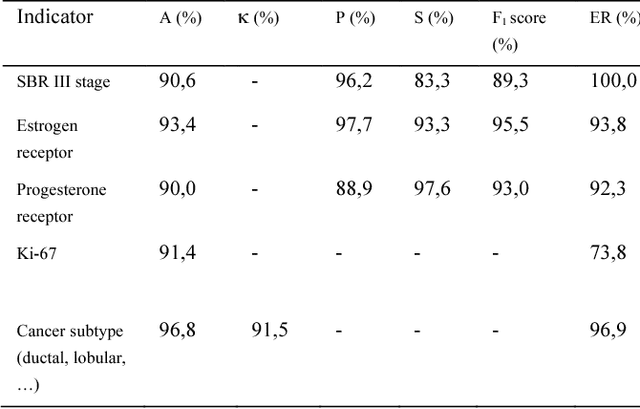
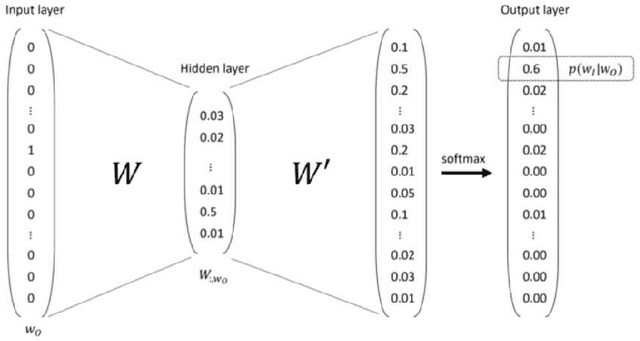
The digitalization of stored information in hospitals now allows for the exploitation of medical data in text format, as electronic health records (EHRs), initially gathered for other purposes than epidemiology. Manual search and analysis operations on such data become tedious. In recent years, the use of natural language processing (NLP) tools was highlighted to automatize the extraction of information contained in EHRs, structure it and perform statistical analysis on this structured information. The main difficulties with the existing approaches is the requirement of synonyms or ontology dictionaries, that are mostly available in English only and do not include local or custom notations. In this work, a team composed of oncologists as domain experts and data scientists develop a custom NLP-based system to process and structure textual clinical reports of patients suffering from breast cancer. The tool relies on the combination of standard text mining techniques and an advanced synonym detection method. It allows for a global analysis by retrieval of indicators such as medical history, tumor characteristics, therapeutic responses, recurrences and prognosis. The versatility of the method allows to obtain easily new indicators, thus opening up the way for retrospective studies with a substantial reduction of the amount of manual work. With no need for biomedical annotators or pre-defined ontologies, this language-agnostic method reached an good extraction accuracy for several concepts of interest, according to a comparison with a manually structured file, without requiring any existing corpus with local or new notations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge