Alex Gorodetsky
A switching Kalman filter approach to online mitigation and correction sensor corruption for inertial navigation
Dec 09, 2024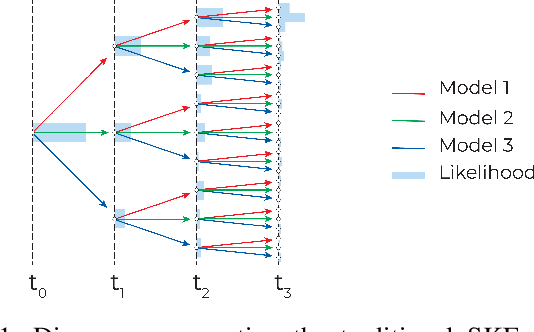
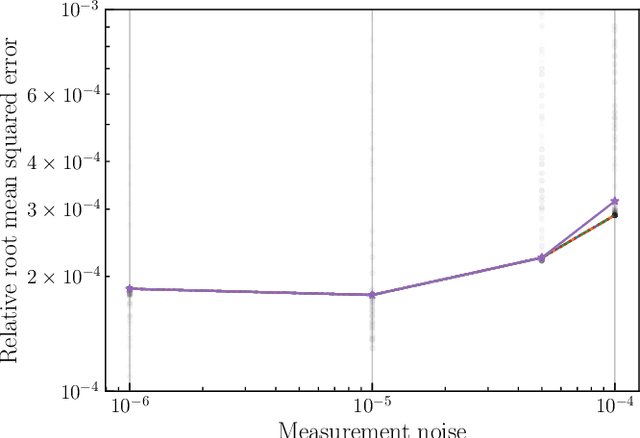
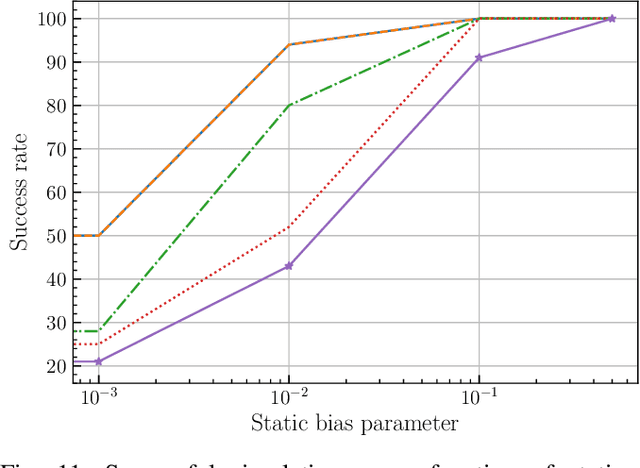
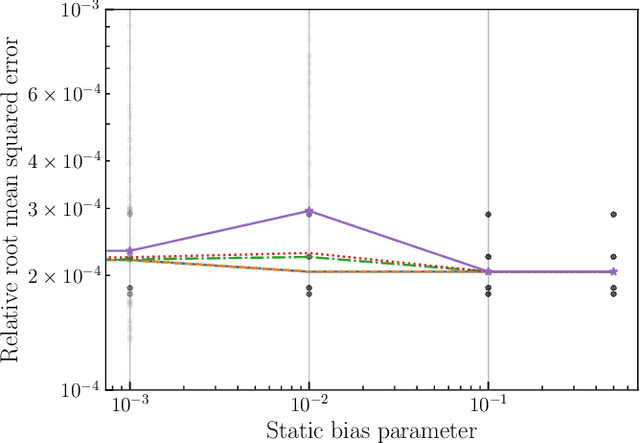
Abstract:This paper introduces a novel approach to detect and address faulty or corrupted external sensors in the context of inertial navigation by leveraging a switching Kalman Filter combined with parameter augmentation. Instead of discarding the corrupted data, the proposed method retains and processes it, running multiple observation models simultaneously and evaluating their likelihoods to accurately identify the true state of the system. We demonstrate the effectiveness of this approach to both identify the moment that a sensor becomes faulty and to correct for the resulting sensor behavior to maintain accurate estimates. We demonstrate our approach on an application of balloon navigation in the atmosphere and shuttle reentry. The results show that our method can accurately recover the true system state even in the presence of significant sensor bias, thereby improving the robustness and reliability of state estimation systems under challenging conditions. We also provide a statistical analysis of problem settings to determine when and where our method is most accurate and where it fails.
Low-rank Bayesian matrix completion via geodesic Hamiltonian Monte Carlo on Stiefel manifolds
Oct 27, 2024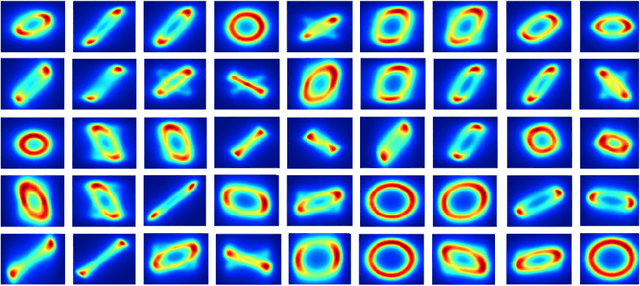
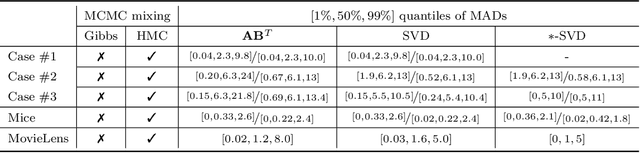
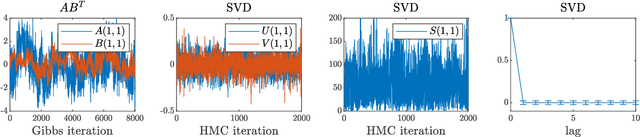
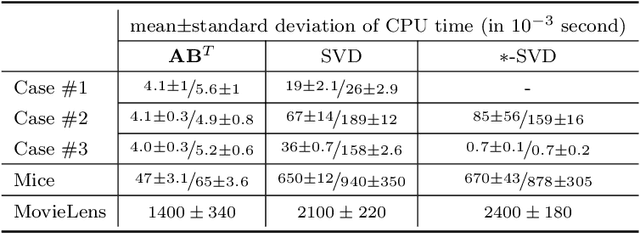
Abstract:We present a new sampling-based approach for enabling efficient computation of low-rank Bayesian matrix completion and quantifying the associated uncertainty. Firstly, we design a new prior model based on the singular-value-decomposition (SVD) parametrization of low-rank matrices. Our prior is analogous to the seminal nuclear-norm regularization used in non-Bayesian setting and enforces orthogonality in the factor matrices by constraining them to Stiefel manifolds. Then, we design a geodesic Hamiltonian Monte Carlo (-within-Gibbs) algorithm for generating posterior samples of the SVD factor matrices. We demonstrate that our approach resolves the sampling difficulties encountered by standard Gibbs samplers for the common two-matrix factorization used in matrix completion. More importantly, the geodesic Hamiltonian sampler allows for sampling in cases with more general likelihoods than the typical Gaussian likelihood and Gaussian prior assumptions adopted in most of the existing Bayesian matrix completion literature. We demonstrate an applications of our approach to fit the categorical data of a mice protein dataset and the MovieLens recommendation problem. Numerical examples demonstrate superior sampling performance, including better mixing and faster convergence to a stationary distribution. Moreover, they demonstrate improved accuracy on the two real-world benchmark problems we considered.
Bioinformatics Retrieval Augmentation Data (BRAD) Digital Assistant
Sep 04, 2024Abstract:We present a prototype for a Bioinformatics Retrieval Augmentation Data (BRAD) digital assistant. BRAD integrates a suite of tools to handle a wide range of bioinformatics tasks, from code execution to online search. We demonstrate BRAD's capabilities through (1) improved question-and-answering with retrieval augmented generation (RAG), (2) BRAD's ability to run and write complex software pipelines, and (3) BRAD's ability to organize and distribute tasks across individual and teams of agents. We use BRAD for automation of bioinformatics workflows, performing tasks ranging from gene enrichment and searching the archive to automatic code generation and running biomarker identification pipelines. BRAD is a step toward the ultimate goal to develop a digital twin of laboratories driven by self-contained loops for hypothesis generation and testing of digital biology experiments.
MFNets: Learning network representations for multifidelity surrogate modeling
Aug 03, 2020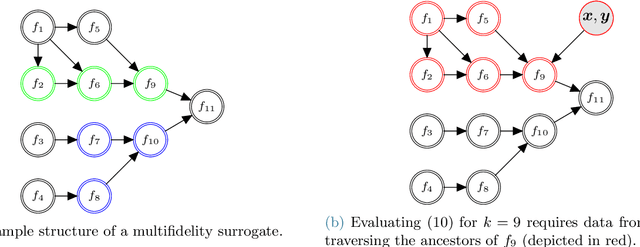
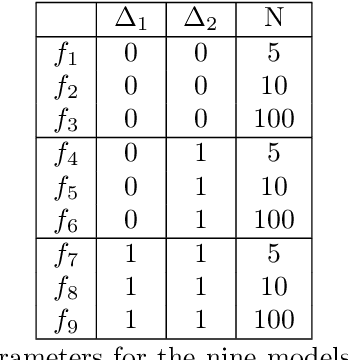
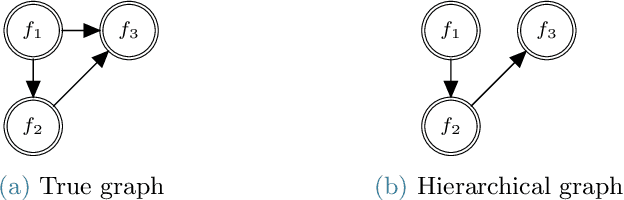
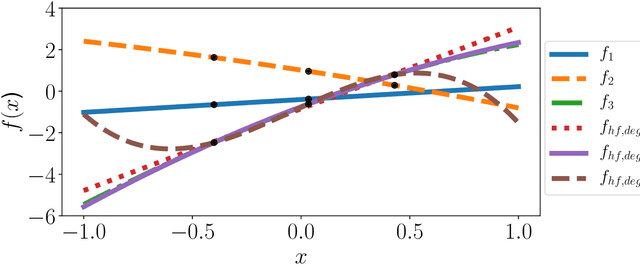
Abstract:This paper presents an approach for constructing multifidelity surrogate models to simultaneously represent, and learn representations of, multiple information sources. The approach formulates a network of surrogate models whose relationships are defined via localized scalings and shifts. The network can have general structure, and can represent a significantly greater variety of modeling relationships than the hierarchical/recursive networks used in the current state of the art. We show empirically that this flexibility achieves greatest gains in the low-data regime, where the network structure must more efficiently leverage the connections between data sources to yield accurate predictions. We demonstrate our approach on four examples ranging from synthetic to physics-based simulation models. For the numerical test cases adopted here, we obtained an order-of-magnitude reduction in errors compared to multifidelity hierarchical and single-fidelity approaches.
Efficient MCMC Sampling for Bayesian Matrix Factorization by Breaking Posterior Symmetries
Jun 08, 2020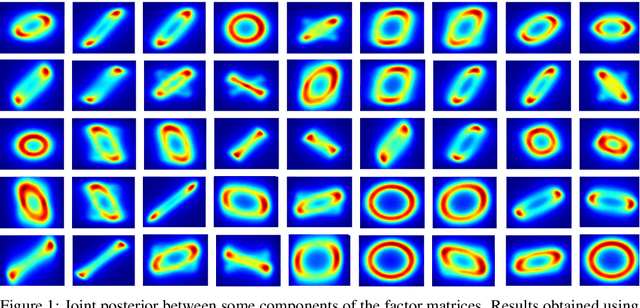
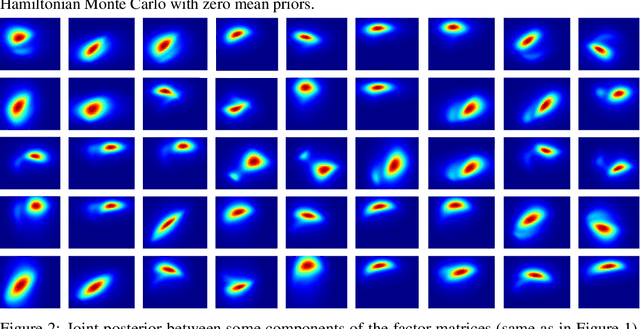
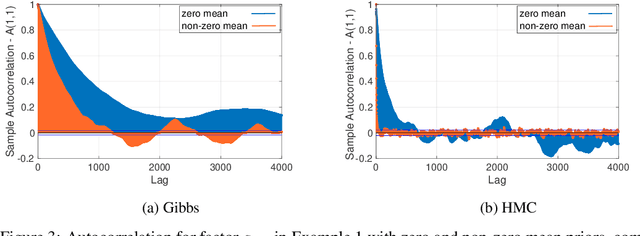
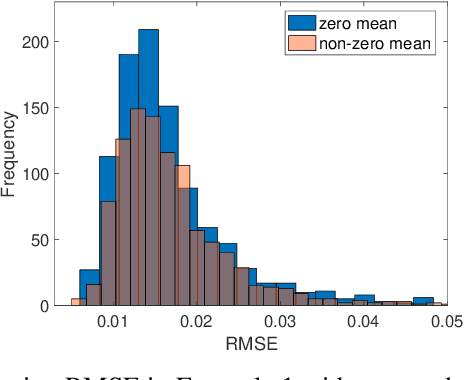
Abstract:Bayesian low-rank matrix factorization techniques have become an essential tool for relational data analysis and matrix completion. A standard approach is to assign zero-mean Gaussian priors on the columns or rows of factor matrices to create a conjugate system. This choice of prior leads to symmetries in the posterior distribution and can severely reduce the efficiency of Markov-chain Monte-Carlo (MCMC) sampling approaches. In this paper, we propose a simple modification to the prior choice that provably breaks these symmetries and maintains/improves accuracy. Specifically, we provide conditions that the Gaussian prior mean and covariance must satisfy so the posterior does not exhibit invariances that yield sampling difficulties. For example, we show that using non-zero linearly independent prior means significantly lowers the autocorrelation of MCMC samples, and can also lead to lower reconstruction errors.
Bayesian System ID: Optimal management of parameter, model, and measurement uncertainty
Mar 04, 2020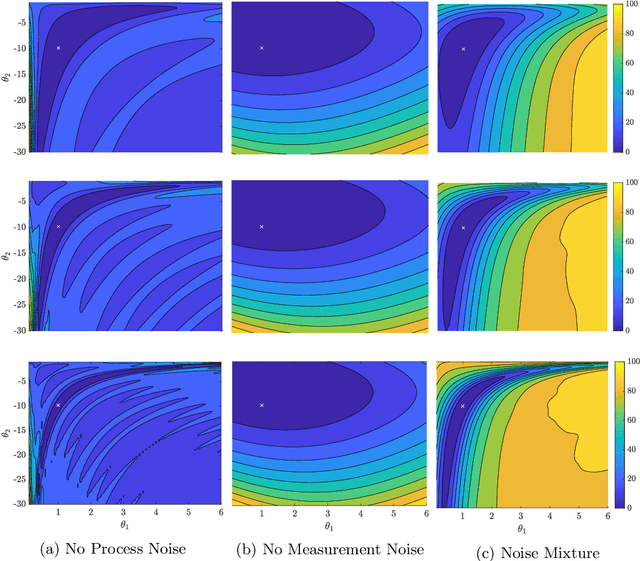
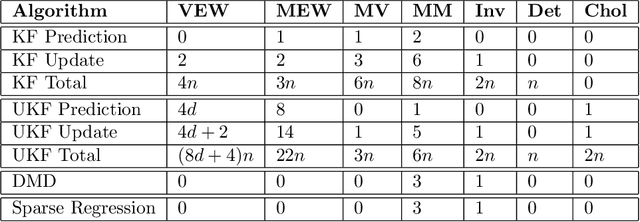
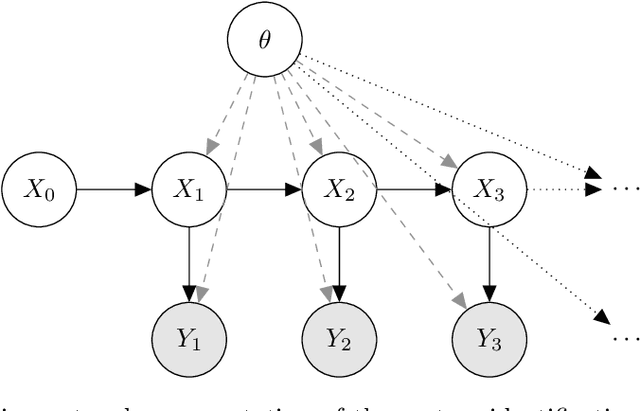
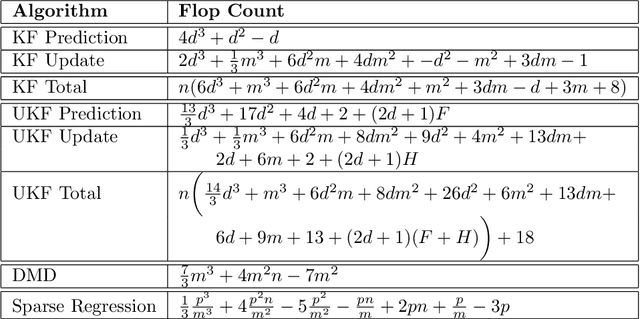
Abstract:We evaluate the robustness of a probabilistic formulation of system identification (ID) to sparse, noisy, and indirect data. Specifically, we compare estimators of future system behavior derived from the Bayesian posterior of a learning problem to several commonly used least squares-based optimization objectives used in system ID. Our comparisons indicate that the log posterior has improved geometric properties compared with the objective function surfaces of traditional methods that include differentially constrained least squares and least squares reconstructions of discrete time steppers like dynamic mode decomposition (DMD). These properties allow it to be both more sensitive to new data and less affected by multiple minima --- overall yielding a more robust approach. Our theoretical results indicate that least squares and regularized least squares methods like dynamic mode decomposition and sparse identification of nonlinear dynamics (SINDy) can be derived from the probabilistic formulation by assuming noiseless measurements. We also analyze the computational complexity of a Gaussian filter-based approximate marginal Markov Chain Monte Carlo scheme that we use to obtain the Bayesian posterior for both linear and nonlinear problems. We then empirically demonstrate that obtaining the marginal posterior of the parameter dynamics and making predictions by extracting optimal estimators (e.g., mean, median, mode) yields orders of magnitude improvement over the aforementioned approaches. We attribute this performance to the fact that the Bayesian approach captures parameter, model, and measurement uncertainties, whereas the other methods typically neglect at least one type of uncertainty.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge