Ziqing Lu
MolCap-Arena: A Comprehensive Captioning Benchmark on Language-Enhanced Molecular Property Prediction
Nov 01, 2024



Abstract:Bridging biomolecular modeling with natural language information, particularly through large language models (LLMs), has recently emerged as a promising interdisciplinary research area. LLMs, having been trained on large corpora of scientific documents, demonstrate significant potential in understanding and reasoning about biomolecules by providing enriched contextual and domain knowledge. However, the extent to which LLM-driven insights can improve performance on complex predictive tasks (e.g., toxicity) remains unclear. Further, the extent to which relevant knowledge can be extracted from LLMs also remains unknown. In this study, we present Molecule Caption Arena: the first comprehensive benchmark of LLM-augmented molecular property prediction. We evaluate over twenty LLMs, including both general-purpose and domain-specific molecule captioners, across diverse prediction tasks. To this goal, we introduce a novel, battle-based rating system. Our findings confirm the ability of LLM-extracted knowledge to enhance state-of-the-art molecular representations, with notable model-, prompt-, and dataset-specific variations. Code, resources, and data are available at github.com/Genentech/molcap-arena.
Cell Morphology-Guided Small Molecule Generation with GFlowNets
Aug 09, 2024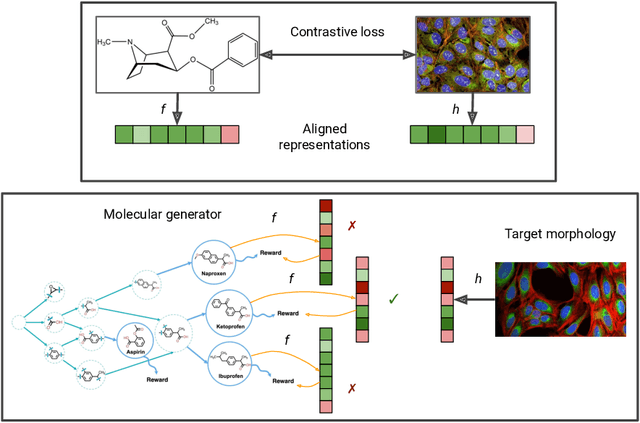

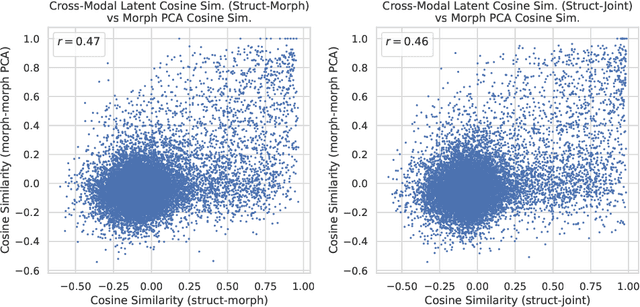

Abstract:High-content phenotypic screening, including high-content imaging (HCI), has gained popularity in the last few years for its ability to characterize novel therapeutics without prior knowledge of the protein target. When combined with deep learning techniques to predict and represent molecular-phenotype interactions, these advancements hold the potential to significantly accelerate and enhance drug discovery applications. This work focuses on the novel task of HCI-guided molecular design. Generative models for molecule design could be guided by HCI data, for example with a supervised model that links molecules to phenotypes of interest as a reward function. However, limited labeled data, combined with the high-dimensional readouts, can make training these methods challenging and impractical. We consider an alternative approach in which we leverage an unsupervised multimodal joint embedding to define a latent similarity as a reward for GFlowNets. The proposed model learns to generate new molecules that could produce phenotypic effects similar to those of the given image target, without relying on pre-annotated phenotypic labels. We demonstrate that the proposed method generates molecules with high morphological and structural similarity to the target, increasing the likelihood of similar biological activity, as confirmed by an independent oracle model.
Optimal Cost Constrained Adversarial Attacks For Multiple Agent Systems
Nov 01, 2023


Abstract:Finding optimal adversarial attack strategies is an important topic in reinforcement learning and the Markov decision process. Previous studies usually assume one all-knowing coordinator (attacker) for whom attacking different recipient (victim) agents incurs uniform costs. However, in reality, instead of using one limitless central attacker, the attacks often need to be performed by distributed attack agents. We formulate the problem of performing optimal adversarial agent-to-agent attacks using distributed attack agents, in which we impose distinct cost constraints on each different attacker-victim pair. We propose an optimal method integrating within-step static constrained attack-resource allocation optimization and between-step dynamic programming to achieve the optimal adversarial attack in a multi-agent system. Our numerical results show that the proposed attacks can significantly reduce the rewards received by the attacked agents.
A 3D-Shape Similarity-based Contrastive Approach to Molecular Representation Learning
Nov 03, 2022Abstract:Molecular shape and geometry dictate key biophysical recognition processes, yet many graph neural networks disregard 3D information for molecular property prediction. Here, we propose a new contrastive-learning procedure for graph neural networks, Molecular Contrastive Learning from Shape Similarity (MolCLaSS), that implicitly learns a three-dimensional representation. Rather than directly encoding or targeting three-dimensional poses, MolCLaSS matches a similarity objective based on Gaussian overlays to learn a meaningful representation of molecular shape. We demonstrate how this framework naturally captures key aspects of three-dimensionality that two-dimensional representations cannot and provides an inductive framework for scaffold hopping.
LocalDrop: A Hybrid Regularization for Deep Neural Networks
Mar 01, 2021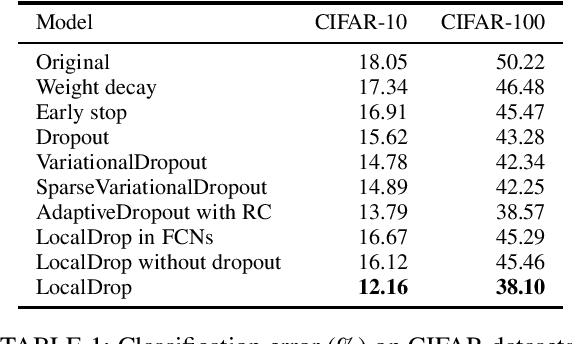
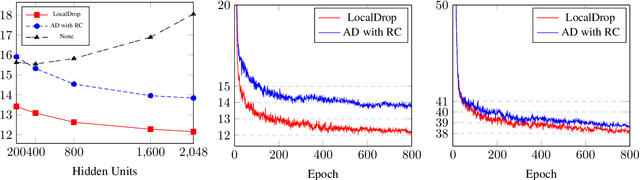
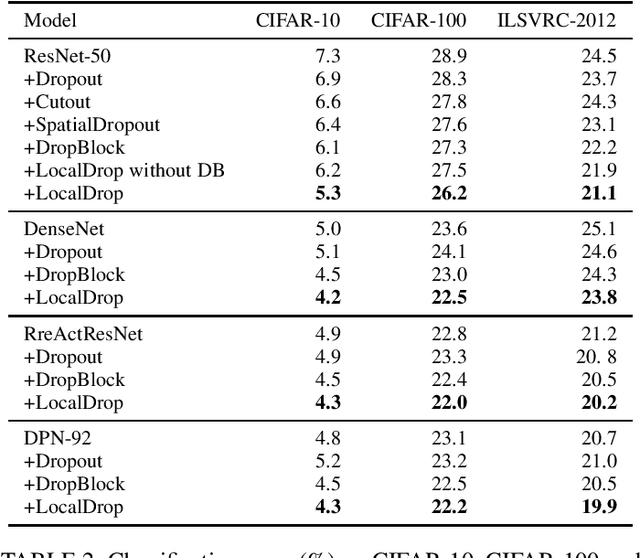
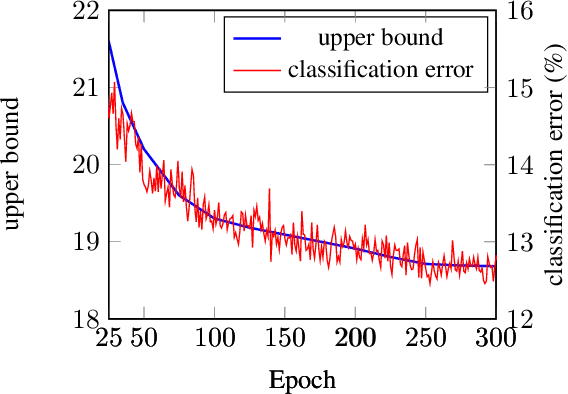
Abstract:In neural networks, developing regularization algorithms to settle overfitting is one of the major study areas. We propose a new approach for the regularization of neural networks by the local Rademacher complexity called LocalDrop. A new regularization function for both fully-connected networks (FCNs) and convolutional neural networks (CNNs), including drop rates and weight matrices, has been developed based on the proposed upper bound of the local Rademacher complexity by the strict mathematical deduction. The analyses of dropout in FCNs and DropBlock in CNNs with keep rate matrices in different layers are also included in the complexity analyses. With the new regularization function, we establish a two-stage procedure to obtain the optimal keep rate matrix and weight matrix to realize the whole training model. Extensive experiments have been conducted to demonstrate the effectiveness of LocalDrop in different models by comparing it with several algorithms and the effects of different hyperparameters on the final performances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge