Zhichao Feng
Context-based Fast Recommendation Strategy for Long User Behavior Sequence in Meituan Waimai
Mar 19, 2024



Abstract:In the recommender system of Meituan Waimai, we are dealing with ever-lengthening user behavior sequences, which pose an increasing challenge to modeling user preference effectively. Existing sequential recommendation models often fail to capture long-term dependencies or are too complex, complicating the fulfillment of Meituan Waimai's unique business needs. To better model user interests, we consider selecting relevant sub-sequences from users' extensive historical behaviors based on their preferences. In this specific scenario, we've noticed that the contexts in which users interact have a significant impact on their preferences. For this purpose, we introduce a novel method called Context-based Fast Recommendation Strategy to tackle the issue of long sequences. We first identify contexts that share similar user preferences with the target context and then locate the corresponding PoIs based on these identified contexts. This approach eliminates the necessity to select a sub-sequence for every candidate PoI, thereby avoiding high time complexity. Specifically, we implement a prototype-based approach to pinpoint contexts that mirror similar user preferences. To amplify accuracy and interpretability, we employ JS divergence of PoI attributes such as categories and prices as a measure of similarity between contexts. A temporal graph integrating both prototype and context nodes helps incorporate temporal information. We then identify appropriate prototypes considering both target contexts and short-term user preferences. Following this, we utilize contexts aligned with these prototypes to generate a sub-sequence, aimed at predicting CTR and CTCVR scores with target attention. Since its inception in 2023, this strategy has been adopted in Meituan Waimai's display recommender system, leading to a 4.6% surge in CTR and a 4.2% boost in GMV.
PACS: Prediction and analysis of cancer subtypes from multi-omics data based on a multi-head attention mechanism model
Aug 21, 2023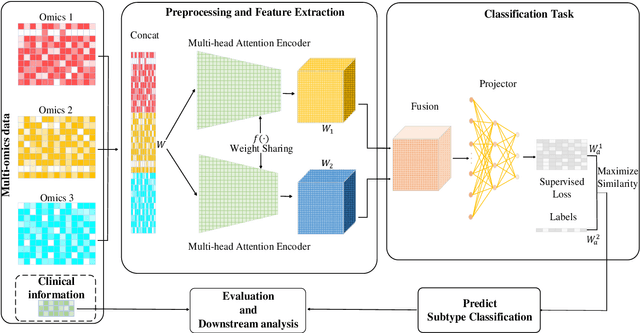
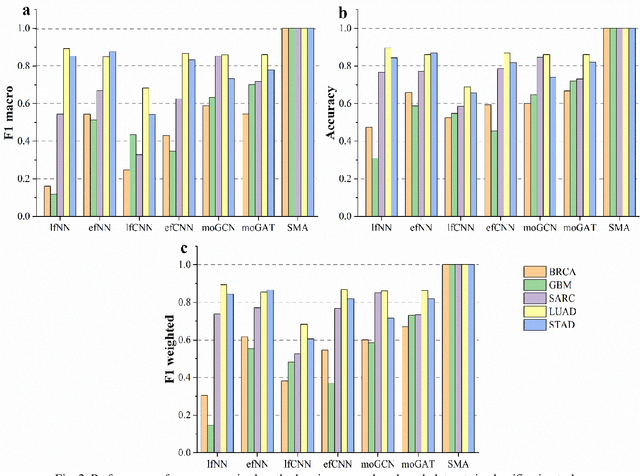
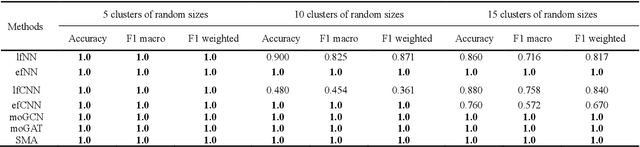
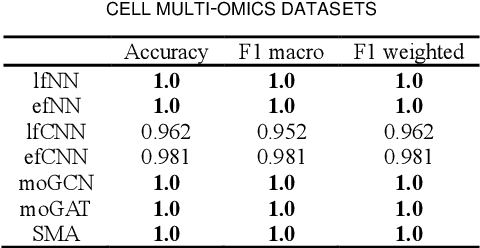
Abstract:Due to the high heterogeneity and clinical characteristics of cancer, there are significant differences in multi-omic data and clinical characteristics among different cancer subtypes. Therefore, accurate classification of cancer subtypes can help doctors choose the most appropriate treatment options, improve treatment outcomes, and provide more accurate patient survival predictions. In this study, we propose a supervised multi-head attention mechanism model (SMA) to classify cancer subtypes successfully. The attention mechanism and feature sharing module of the SMA model can successfully learn the global and local feature information of multi-omics data. Second, it enriches the parameters of the model by deeply fusing multi-head attention encoders from Siamese through the fusion module. Validated by extensive experiments, the SMA model achieves the highest accuracy, F1 macroscopic, F1 weighted, and accurate classification of cancer subtypes in simulated, single-cell, and cancer multiomics datasets compared to AE, CNN, and GNN-based models. Therefore, we contribute to future research on multiomics data using our attention-based approach.
CVFC: Attention-Based Cross-View Feature Consistency for Weakly Supervised Semantic Segmentation of Pathology Images
Aug 21, 2023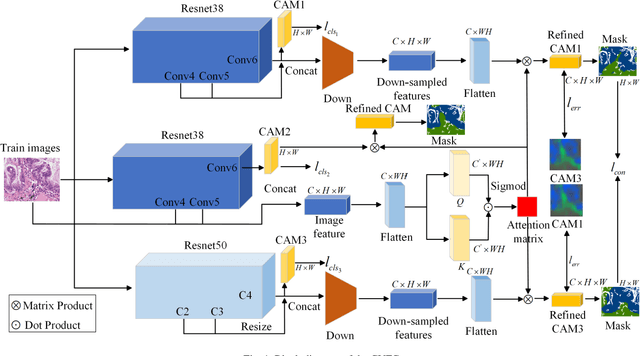
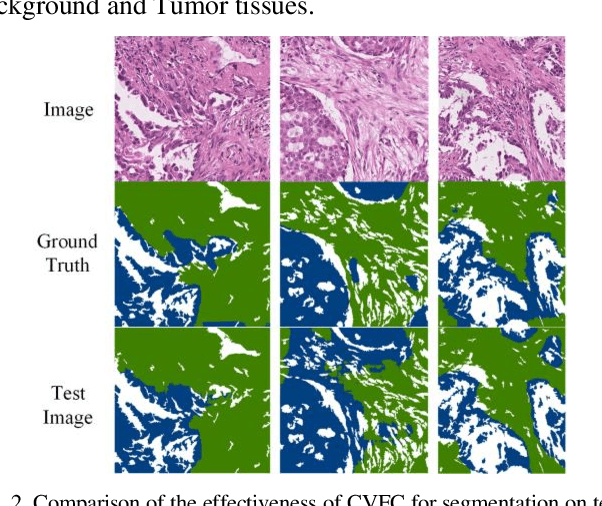
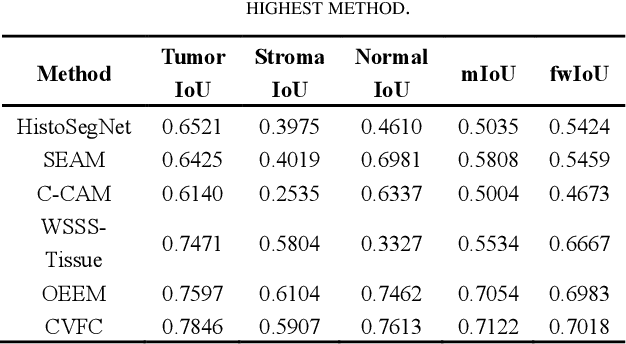
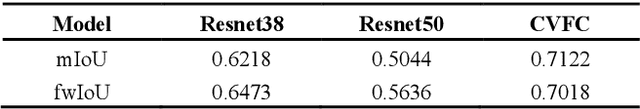
Abstract:Histopathology image segmentation is the gold standard for diagnosing cancer, and can indicate cancer prognosis. However, histopathology image segmentation requires high-quality masks, so many studies now use imagelevel labels to achieve pixel-level segmentation to reduce the need for fine-grained annotation. To solve this problem, we propose an attention-based cross-view feature consistency end-to-end pseudo-mask generation framework named CVFC based on the attention mechanism. Specifically, CVFC is a three-branch joint framework composed of two Resnet38 and one Resnet50, and the independent branch multi-scale integrated feature map to generate a class activation map (CAM); in each branch, through down-sampling and The expansion method adjusts the size of the CAM; the middle branch projects the feature matrix to the query and key feature spaces, and generates a feature space perception matrix through the connection layer and inner product to adjust and refine the CAM of each branch; finally, through the feature consistency loss and feature cross loss to optimize the parameters of CVFC in co-training mode. After a large number of experiments, An IoU of 0.7122 and a fwIoU of 0.7018 are obtained on the WSSS4LUAD dataset, which outperforms HistoSegNet, SEAM, C-CAM, WSSS-Tissue, and OEEM, respectively.
LDCSF: Local depth convolution-based Swim framework for classifying multi-label histopathology images
Aug 21, 2023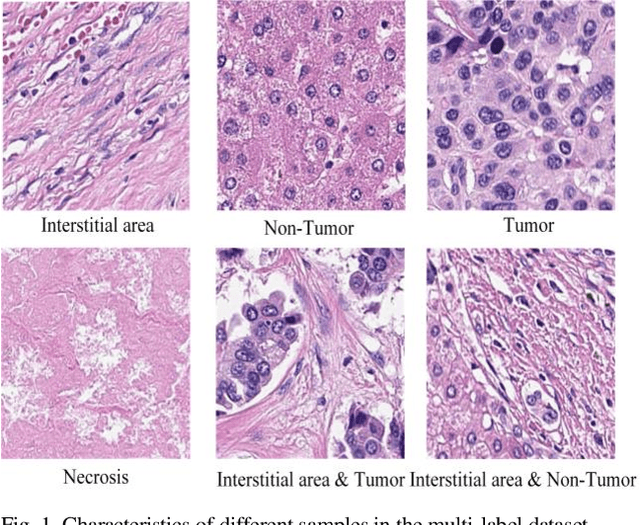
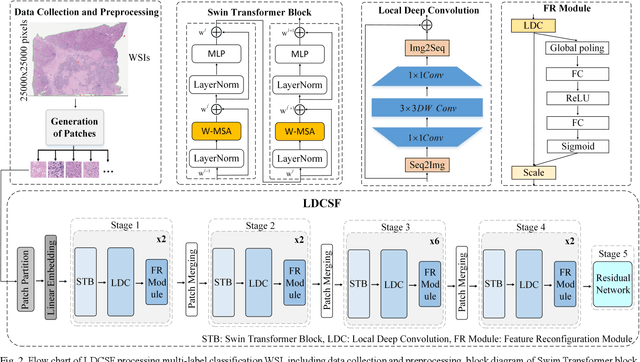
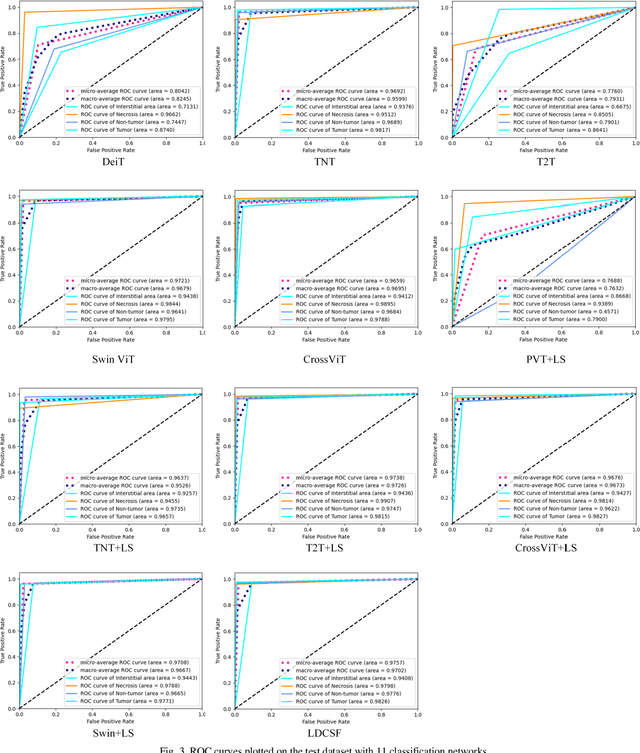
Abstract:Histopathological images are the gold standard for diagnosing liver cancer. However, the accuracy of fully digital diagnosis in computational pathology needs to be improved. In this paper, in order to solve the problem of multi-label and low classification accuracy of histopathology images, we propose a locally deep convolutional Swim framework (LDCSF) to classify multi-label histopathology images. In order to be able to provide local field of view diagnostic results, we propose the LDCSF model, which consists of a Swin transformer module, a local depth convolution (LDC) module, a feature reconstruction (FR) module, and a ResNet module. The Swin transformer module reduces the amount of computation generated by the attention mechanism by limiting the attention to each window. The LDC then reconstructs the attention map and performs convolution operations in multiple channels, passing the resulting feature map to the next layer. The FR module uses the corresponding weight coefficient vectors obtained from the channels to dot product with the original feature map vector matrix to generate representative feature maps. Finally, the residual network undertakes the final classification task. As a result, the classification accuracy of LDCSF for interstitial area, necrosis, non-tumor and tumor reached 0.9460, 0.9960, 0.9808, 0.9847, respectively. Finally, we use the results of multi-label pathological image classification to calculate the tumor-to-stromal ratio, which lays the foundation for the analysis of the microenvironment of liver cancer histopathological images. Second, we released a multilabel histopathology image of liver cancer, our code and data are available at https://github.com/panliangrui/LSF.
Heterogeneous Knowledge Fusion: A Novel Approach for Personalized Recommendation via LLM
Aug 18, 2023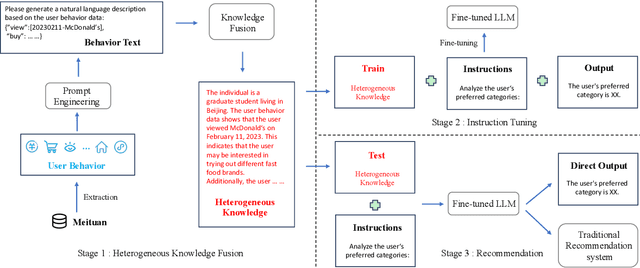
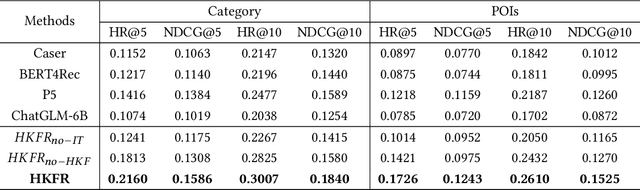
Abstract:The analysis and mining of user heterogeneous behavior are of paramount importance in recommendation systems. However, the conventional approach of incorporating various types of heterogeneous behavior into recommendation models leads to feature sparsity and knowledge fragmentation issues. To address this challenge, we propose a novel approach for personalized recommendation via Large Language Model (LLM), by extracting and fusing heterogeneous knowledge from user heterogeneous behavior information. In addition, by combining heterogeneous knowledge and recommendation tasks, instruction tuning is performed on LLM for personalized recommendations. The experimental results demonstrate that our method can effectively integrate user heterogeneous behavior and significantly improve recommendation performance.
Multi-Head Attention Mechanism Learning for Cancer New Subtypes and Treatment Based on Cancer Multi-Omics Data
Jul 09, 2023Abstract:Due to the high heterogeneity and clinical characteristics of cancer, there are significant differences in multi-omics data and clinical features among subtypes of different cancers. Therefore, the identification and discovery of cancer subtypes are crucial for the diagnosis, treatment, and prognosis of cancer. In this study, we proposed a generalization framework based on attention mechanisms for unsupervised contrastive learning (AMUCL) to analyze cancer multi-omics data for the identification and characterization of cancer subtypes. AMUCL framework includes a unsupervised multi-head attention mechanism, which deeply extracts multi-omics data features. Importantly, a decoupled contrastive learning model (DMACL) based on a multi-head attention mechanism is proposed to learn multi-omics data features and clusters and identify new cancer subtypes. This unsupervised contrastive learning method clusters subtypes by calculating the similarity between samples in the feature space and sample space of multi-omics data. Compared to 11 other deep learning models, the DMACL model achieved a C-index of 0.002, a Silhouette score of 0.801, and a Davies Bouldin Score of 0.38 on a single-cell multi-omics dataset. On a cancer multi-omics dataset, the DMACL model obtained a C-index of 0.016, a Silhouette score of 0.688, and a Davies Bouldin Score of 0.46, and obtained the most reliable cancer subtype clustering results for each type of cancer. Finally, we used the DMACL model in the AMUCL framework to reveal six cancer subtypes of AML. By analyzing the GO functional enrichment, subtype-specific biological functions, and GSEA of AML, we further enhanced the interpretability of cancer subtype analysis based on the generalizable AMUCL framework.
A review of machine learning approaches, challenges and prospects for computational tumor pathology
May 31, 2022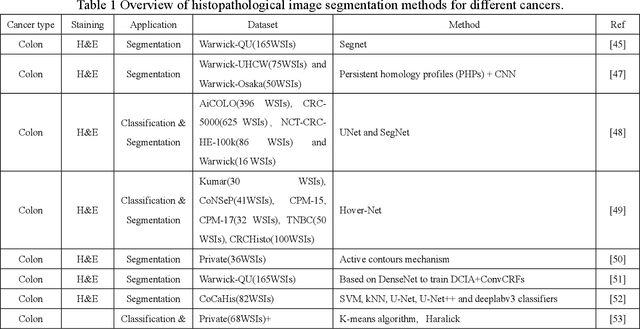

Abstract:Computational pathology is part of precision oncology medicine. The integration of high-throughput data including genomics, transcriptomics, proteomics, metabolomics, pathomics, and radiomics into clinical practice improves cancer treatment plans, treatment cycles, and cure rates, and helps doctors open up innovative approaches to patient prognosis. In the past decade, rapid advances in artificial intelligence, chip design and manufacturing, and mobile computing have facilitated research in computational pathology and have the potential to provide better-integrated solutions for whole-slide images, multi-omics data, and clinical informatics. However, tumor computational pathology now brings some challenges to the application of tumour screening, diagnosis and prognosis in terms of data integration, hardware processing, network sharing bandwidth and machine learning technology. This review investigates image preprocessing methods in computational pathology from a pathological and technical perspective, machine learning-based methods, and applications of computational pathology in breast, colon, prostate, lung, and various tumour disease scenarios. Finally, the challenges and prospects of machine learning in computational pathology applications are discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge