Lian Wang
FedDP: Privacy-preserving method based on federated learning for histopathology image segmentation
Nov 07, 2024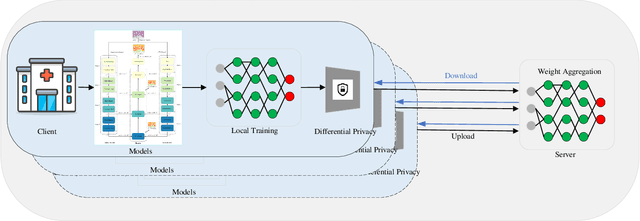
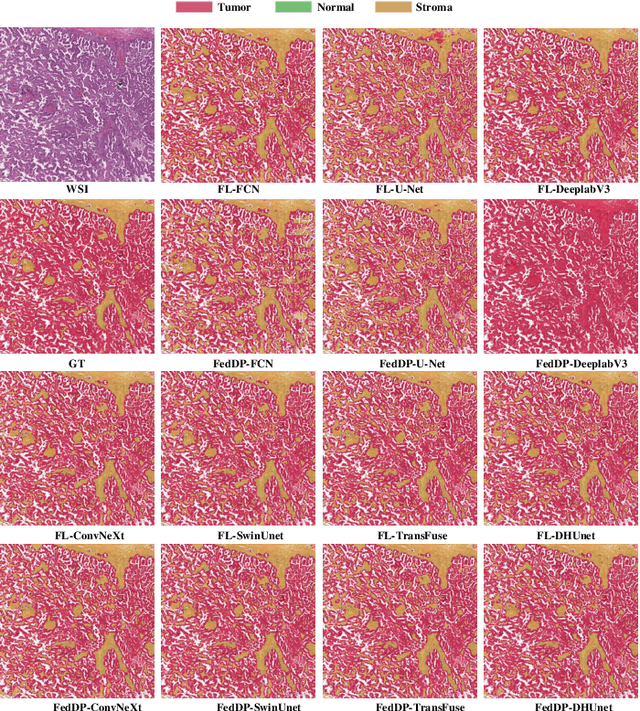
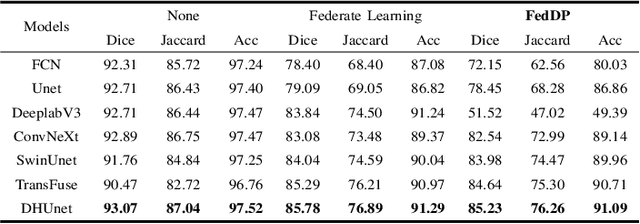
Abstract:Hematoxylin and Eosin (H&E) staining of whole slide images (WSIs) is considered the gold standard for pathologists and medical practitioners for tumor diagnosis, surgical planning, and post-operative assessment. With the rapid advancement of deep learning technologies, the development of numerous models based on convolutional neural networks and transformer-based models has been applied to the precise segmentation of WSIs. However, due to privacy regulations and the need to protect patient confidentiality, centralized storage and processing of image data are impractical. Training a centralized model directly is challenging to implement in medical settings due to these privacy concerns.This paper addresses the dispersed nature and privacy sensitivity of medical image data by employing a federated learning framework, allowing medical institutions to collaboratively learn while protecting patient privacy. Additionally, to address the issue of original data reconstruction through gradient inversion during the federated learning training process, differential privacy introduces noise into the model updates, preventing attackers from inferring the contributions of individual samples, thereby protecting the privacy of the training data.Experimental results show that the proposed method, FedDP, minimally impacts model accuracy while effectively safeguarding the privacy of cancer pathology image data, with only a slight decrease in Dice, Jaccard, and Acc indices by 0.55%, 0.63%, and 0.42%, respectively. This approach facilitates cross-institutional collaboration and knowledge sharing while protecting sensitive data privacy, providing a viable solution for further research and application in the medical field.
CVFC: Attention-Based Cross-View Feature Consistency for Weakly Supervised Semantic Segmentation of Pathology Images
Aug 21, 2023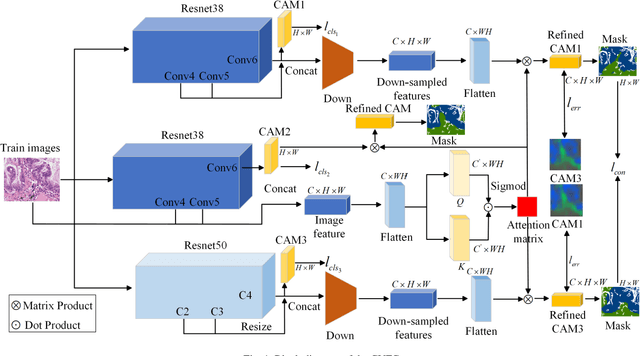
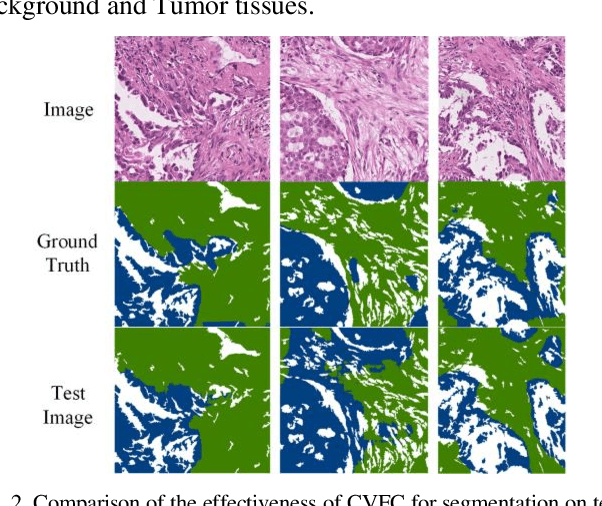
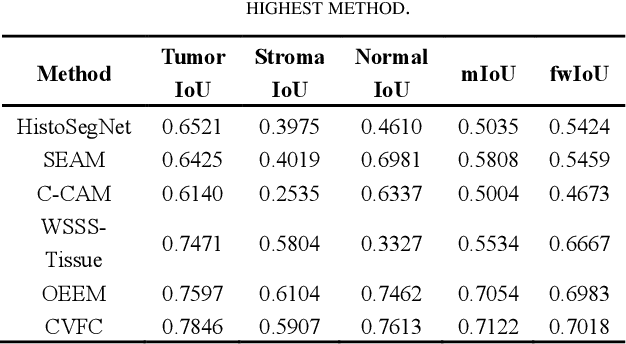
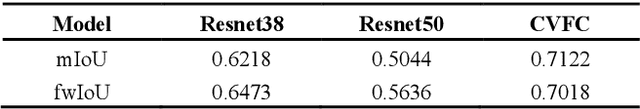
Abstract:Histopathology image segmentation is the gold standard for diagnosing cancer, and can indicate cancer prognosis. However, histopathology image segmentation requires high-quality masks, so many studies now use imagelevel labels to achieve pixel-level segmentation to reduce the need for fine-grained annotation. To solve this problem, we propose an attention-based cross-view feature consistency end-to-end pseudo-mask generation framework named CVFC based on the attention mechanism. Specifically, CVFC is a three-branch joint framework composed of two Resnet38 and one Resnet50, and the independent branch multi-scale integrated feature map to generate a class activation map (CAM); in each branch, through down-sampling and The expansion method adjusts the size of the CAM; the middle branch projects the feature matrix to the query and key feature spaces, and generates a feature space perception matrix through the connection layer and inner product to adjust and refine the CAM of each branch; finally, through the feature consistency loss and feature cross loss to optimize the parameters of CVFC in co-training mode. After a large number of experiments, An IoU of 0.7122 and a fwIoU of 0.7018 are obtained on the WSSS4LUAD dataset, which outperforms HistoSegNet, SEAM, C-CAM, WSSS-Tissue, and OEEM, respectively.
Multi-Head Attention Mechanism Learning for Cancer New Subtypes and Treatment Based on Cancer Multi-Omics Data
Jul 09, 2023Abstract:Due to the high heterogeneity and clinical characteristics of cancer, there are significant differences in multi-omics data and clinical features among subtypes of different cancers. Therefore, the identification and discovery of cancer subtypes are crucial for the diagnosis, treatment, and prognosis of cancer. In this study, we proposed a generalization framework based on attention mechanisms for unsupervised contrastive learning (AMUCL) to analyze cancer multi-omics data for the identification and characterization of cancer subtypes. AMUCL framework includes a unsupervised multi-head attention mechanism, which deeply extracts multi-omics data features. Importantly, a decoupled contrastive learning model (DMACL) based on a multi-head attention mechanism is proposed to learn multi-omics data features and clusters and identify new cancer subtypes. This unsupervised contrastive learning method clusters subtypes by calculating the similarity between samples in the feature space and sample space of multi-omics data. Compared to 11 other deep learning models, the DMACL model achieved a C-index of 0.002, a Silhouette score of 0.801, and a Davies Bouldin Score of 0.38 on a single-cell multi-omics dataset. On a cancer multi-omics dataset, the DMACL model obtained a C-index of 0.016, a Silhouette score of 0.688, and a Davies Bouldin Score of 0.46, and obtained the most reliable cancer subtype clustering results for each type of cancer. Finally, we used the DMACL model in the AMUCL framework to reveal six cancer subtypes of AML. By analyzing the GO functional enrichment, subtype-specific biological functions, and GSEA of AML, we further enhanced the interpretability of cancer subtype analysis based on the generalizable AMUCL framework.
Noise-reducing attention cross fusion learning transformer for histological image classification of osteosarcoma
Apr 29, 2022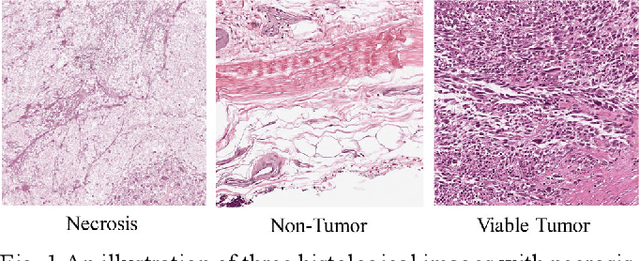
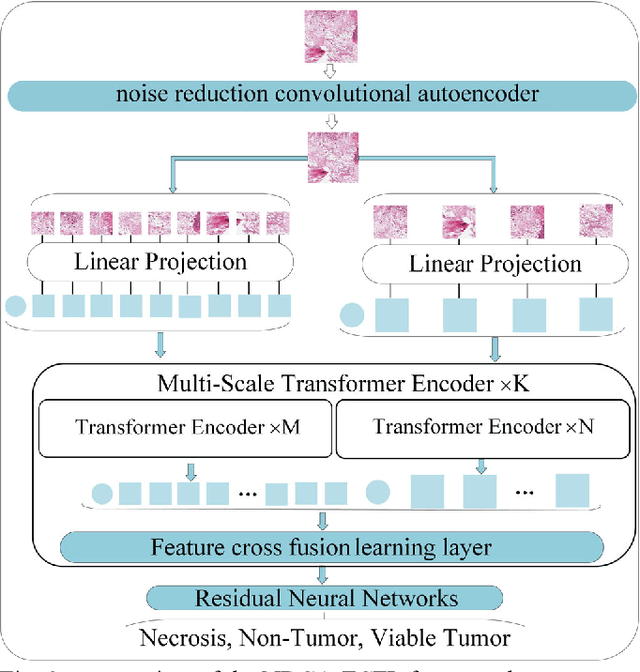
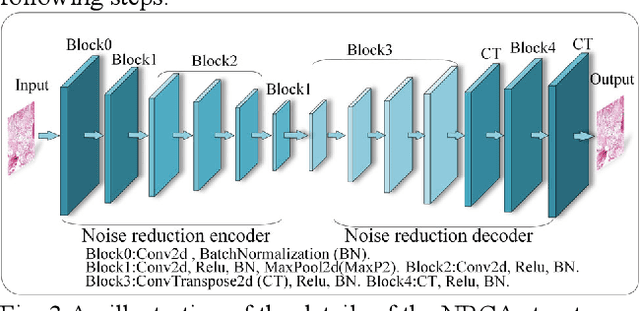
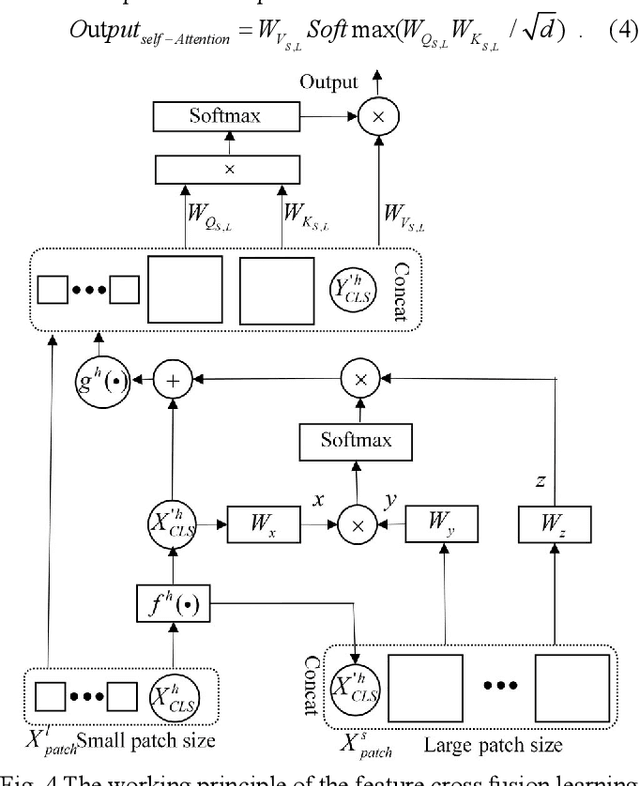
Abstract:The degree of malignancy of osteosarcoma and its tendency to metastasize/spread mainly depend on the pathological grade (determined by observing the morphology of the tumor under a microscope). The purpose of this study is to use artificial intelligence to classify osteosarcoma histological images and to assess tumor survival and necrosis, which will help doctors reduce their workload, improve the accuracy of osteosarcoma cancer detection, and make a better prognosis for patients. The study proposes a typical transformer image classification framework by integrating noise reduction convolutional autoencoder and feature cross fusion learning (NRCA-FCFL) to classify osteosarcoma histological images. Noise reduction convolutional autoencoder could well denoise histological images of osteosarcoma, resulting in more pure images for osteosarcoma classification. Moreover, we introduce feature cross fusion learning, which integrates two scale image patches, to sufficiently explore their interactions by using additional classification tokens. As a result, a refined fusion feature is generated, which is fed to the residual neural network for label predictions. We conduct extensive experiments to evaluate the performance of the proposed approach. The experimental results demonstrate that our method outperforms the traditional and deep learning approaches on various evaluation metrics, with an accuracy of 99.17% to support osteosarcoma diagnosis.
Referring Expression Comprehension via Cross-Level Multi-Modal Fusion
Apr 21, 2022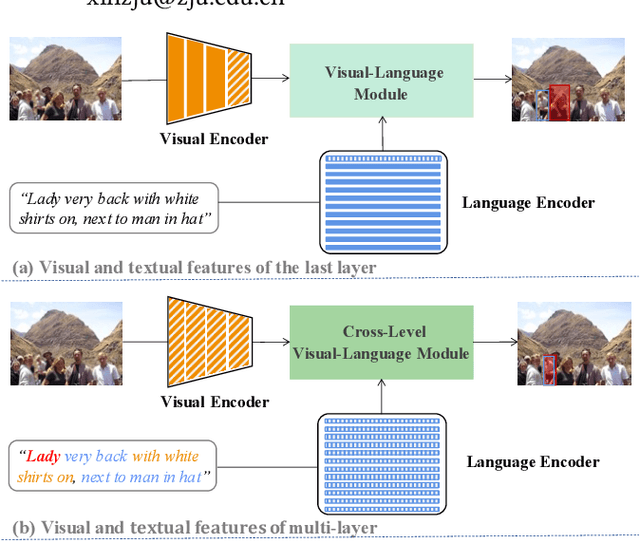
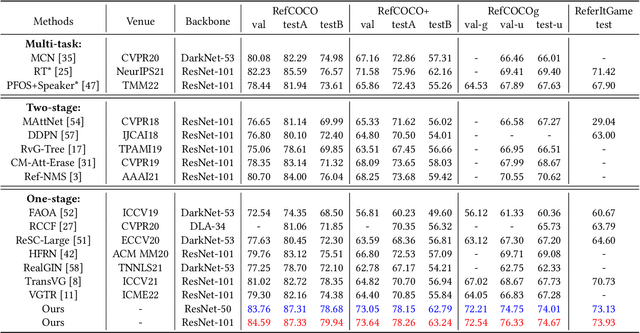

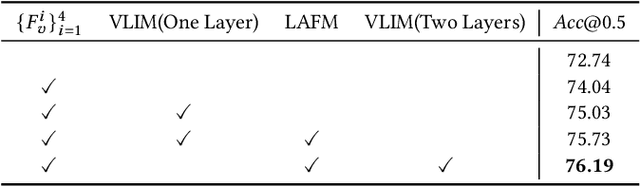
Abstract:As an important and challenging problem in vision-language tasks, referring expression comprehension (REC) aims to localize the target object specified by a given referring expression. Recently, most of the state-of-the-art REC methods mainly focus on multi-modal fusion while overlooking the inherent hierarchical information contained in visual and language encoders. Considering that REC requires visual and textual hierarchical information for accurate target localization, and encoders inherently extract features in a hierarchical fashion, we propose to effectively utilize the rich hierarchical information contained in different layers of visual and language encoders. To this end, we design a Cross-level Multi-modal Fusion (CMF) framework, which gradually integrates visual and textual features of multi-layer through intra- and inter-modal. Experimental results on RefCOCO, RefCOCO+, RefCOCOg, and ReferItGame datasets demonstrate the proposed framework achieves significant performance improvements over state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge