Yuxuan Zheng
Bi-level Mean Field: Dynamic Grouping for Large-Scale MARL
May 10, 2025Abstract:Large-scale Multi-Agent Reinforcement Learning (MARL) often suffers from the curse of dimensionality, as the exponential growth in agent interactions significantly increases computational complexity and impedes learning efficiency. To mitigate this, existing efforts that rely on Mean Field (MF) simplify the interaction landscape by approximating neighboring agents as a single mean agent, thus reducing overall complexity to pairwise interactions. However, these MF methods inevitably fail to account for individual differences, leading to aggregation noise caused by inaccurate iterative updates during MF learning. In this paper, we propose a Bi-level Mean Field (BMF) method to capture agent diversity with dynamic grouping in large-scale MARL, which can alleviate aggregation noise via bi-level interaction. Specifically, BMF introduces a dynamic group assignment module, which employs a Variational AutoEncoder (VAE) to learn the representations of agents, facilitating their dynamic grouping over time. Furthermore, we propose a bi-level interaction module to model both inter- and intra-group interactions for effective neighboring aggregation. Experiments across various tasks demonstrate that the proposed BMF yields results superior to the state-of-the-art methods. Our code will be made publicly available.
Odyssey: Empowering Agents with Open-World Skills
Jul 22, 2024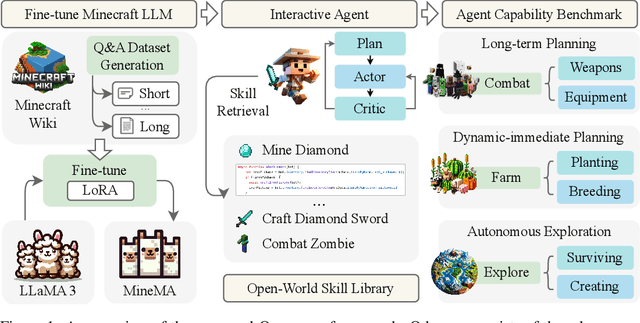
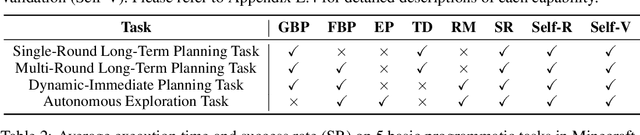
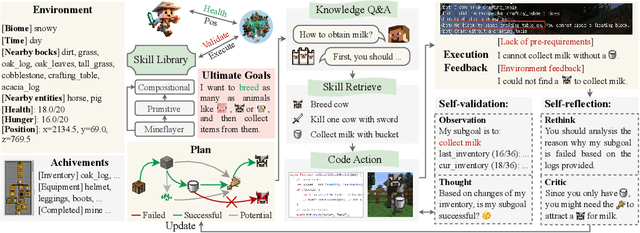
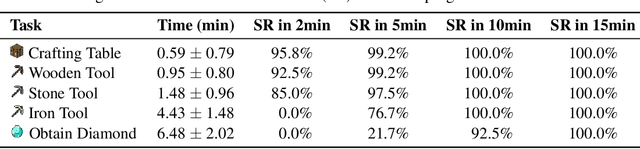
Abstract:Recent studies have delved into constructing generalist agents for open-world embodied environments like Minecraft. Despite the encouraging results, existing efforts mainly focus on solving basic programmatic tasks, e.g., material collection and tool-crafting following the Minecraft tech-tree, treating the ObtainDiamond task as the ultimate goal. This limitation stems from the narrowly defined set of actions available to agents, requiring them to learn effective long-horizon strategies from scratch. Consequently, discovering diverse gameplay opportunities in the open world becomes challenging. In this work, we introduce ODYSSEY, a new framework that empowers Large Language Model (LLM)-based agents with open-world skills to explore the vast Minecraft world. ODYSSEY comprises three key parts: (1) An interactive agent with an open-world skill library that consists of 40 primitive skills and 183 compositional skills. (2) A fine-tuned LLaMA-3 model trained on a large question-answering dataset with 390k+ instruction entries derived from the Minecraft Wiki. (3) A new open-world benchmark includes thousands of long-term planning tasks, tens of dynamic-immediate planning tasks, and one autonomous exploration task. Extensive experiments demonstrate that the proposed ODYSSEY framework can effectively evaluate the planning and exploration capabilities of agents. All datasets, model weights, and code are publicly available to motivate future research on more advanced autonomous agent solutions.
Multi-modal Self-supervised Pre-training for Regulatory Genome Across Cell Types
Nov 03, 2021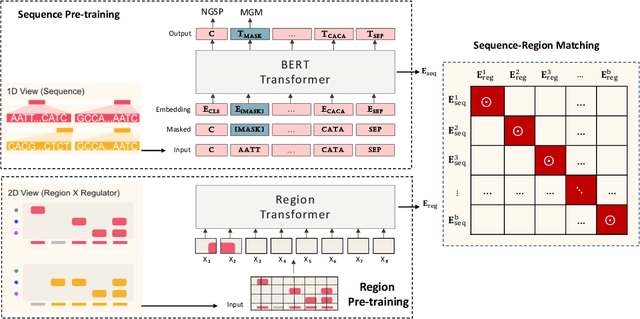

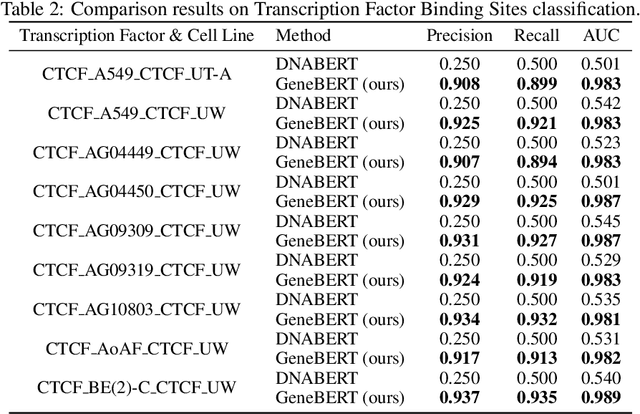
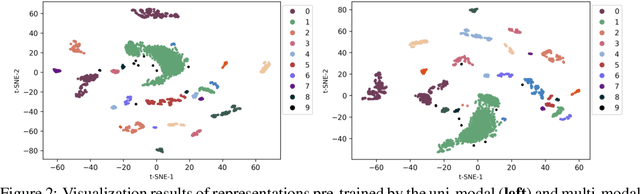
Abstract:In the genome biology research, regulatory genome modeling is an important topic for many regulatory downstream tasks, such as promoter classification, transaction factor binding sites prediction. The core problem is to model how regulatory elements interact with each other and its variability across different cell types. However, current deep learning methods often focus on modeling genome sequences of a fixed set of cell types and do not account for the interaction between multiple regulatory elements, making them only perform well on the cell types in the training set and lack the generalizability required in biological applications. In this work, we propose a simple yet effective approach for pre-training genome data in a multi-modal and self-supervised manner, which we call GeneBERT. Specifically, we simultaneously take the 1d sequence of genome data and a 2d matrix of (transcription factors x regions) as the input, where three pre-training tasks are proposed to improve the robustness and generalizability of our model. We pre-train our model on the ATAC-seq dataset with 17 million genome sequences. We evaluate our GeneBERT on regulatory downstream tasks across different cell types, including promoter classification, transaction factor binding sites prediction, disease risk estimation, and splicing sites prediction. Extensive experiments demonstrate the effectiveness of multi-modal and self-supervised pre-training for large-scale regulatory genomics data.
Horizontally Fused Training Array: An Effective Hardware Utilization Squeezer for Training Novel Deep Learning Models
Feb 07, 2021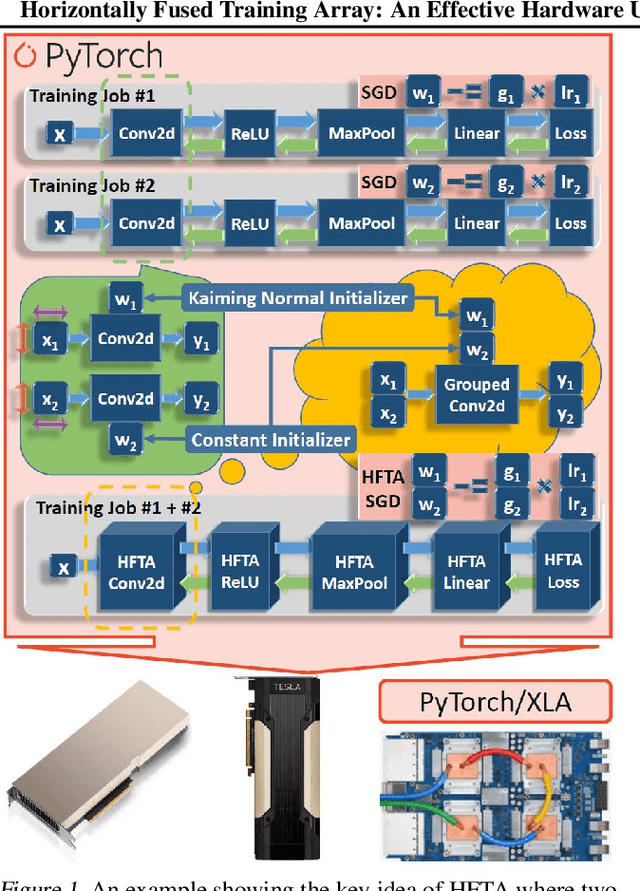
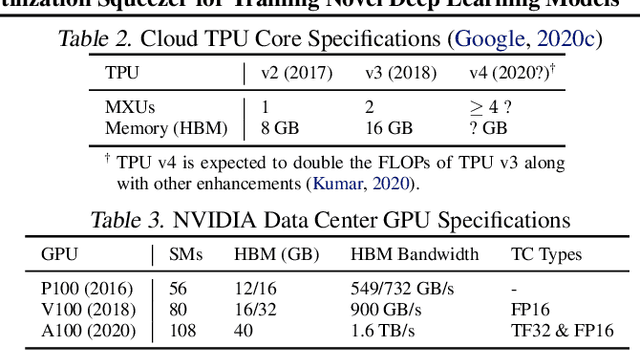

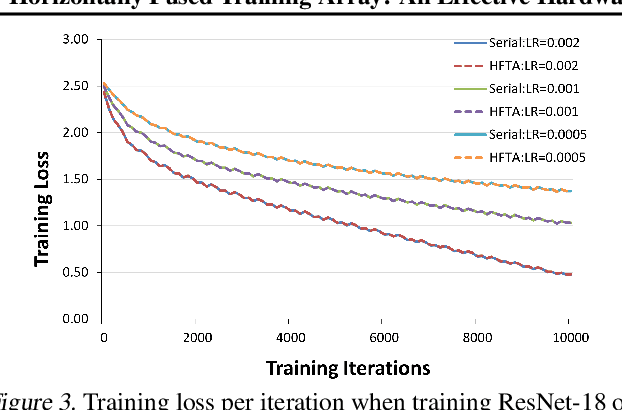
Abstract:Driven by the tremendous effort in researching novel deep learning (DL) algorithms, the training cost of developing new models increases staggeringly in recent years. To reduce this training cost and optimize the cluster-wide hardware resource usage, we analyze GPU cluster usage statistics from a well-known research institute. Our study reveals that single-accelerator training jobs can dominate the cluster-wide resource consumption when launched repetitively (e.g., for hyper-parameter tuning) while severely underutilizing the hardware. This is because DL researchers and practitioners often lack the required expertise to independently optimize their own workloads. Fortunately, we observe that such workloads have the following unique characteristics: (i) the models among jobs often have the same types of operators with the same shapes, and (ii) the inter-model horizontal fusion of such operators is mathematically equivalent to other already well-optimized operators. Thus, to help DL researchers and practitioners effectively and easily improve the hardware utilization of their novel DL training workloads, we propose Horizontally Fused Training Array (HFTA). HFTA is a new DL framework extension library that horizontally fuses the models from different repetitive jobs deeply down to operators, and then trains those models simultaneously on a shared accelerator. On three emerging DL training workloads and state-of-the-art accelerators (GPUs and TPUs), HFTA demonstrates strong effectiveness in squeezing out hardware utilization and achieves up to $15.1 \times$ higher training throughput vs. the standard practice of running each job on a separate accelerator.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge