Yuk-Lam Ho
Representation Learning to Advance Multi-institutional Studies with Electronic Health Record Data
Feb 12, 2025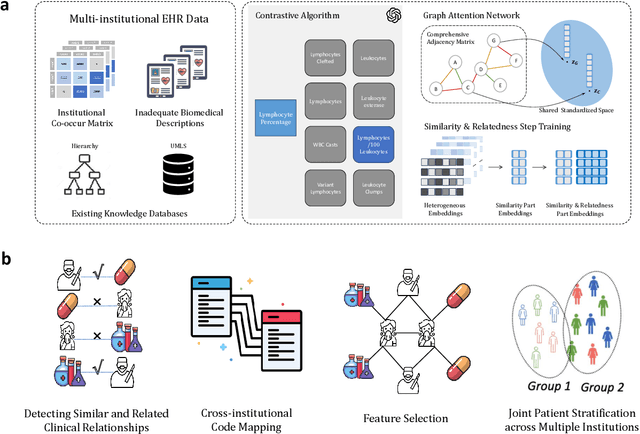
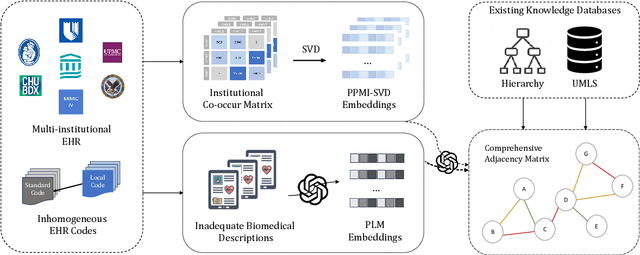
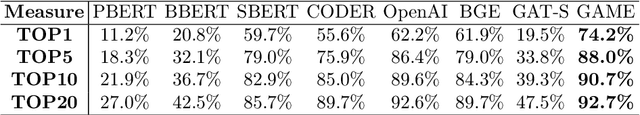
Abstract:The adoption of EHRs has expanded opportunities to leverage data-driven algorithms in clinical care and research. A major bottleneck in effectively conducting multi-institutional EHR studies is the data heterogeneity across systems with numerous codes that either do not exist or represent different clinical concepts across institutions. The need for data privacy further limits the feasibility of including multi-institutional patient-level data required to study similarities and differences across patient subgroups. To address these challenges, we developed the GAME algorithm. Tested and validated across 7 institutions and 2 languages, GAME integrates data in several levels: (1) at the institutional level with knowledge graphs to establish relationships between codes and existing knowledge sources, providing the medical context for standard codes and their relationship to each other; (2) between institutions, leveraging language models to determine the relationships between institution-specific codes with established standard codes; and (3) quantifying the strength of the relationships between codes using a graph attention network. Jointly trained embeddings are created using transfer and federated learning to preserve data privacy. In this study, we demonstrate the applicability of GAME in selecting relevant features as inputs for AI-driven algorithms in a range of conditions, e.g., heart failure, rheumatoid arthritis. We then highlight the application of GAME harmonized multi-institutional EHR data in a study of Alzheimer's disease outcomes and suicide risk among patients with mental health disorders, without sharing patient-level data outside individual institutions.
Domain Shift Analysis in Chest Radiographs Classification in a Veterans Healthcare Administration Population
Jul 30, 2024



Abstract:Objectives: This study aims to assess the impact of domain shift on chest X-ray classification accuracy and to analyze the influence of ground truth label quality and demographic factors such as age group, sex, and study year. Materials and Methods: We used a DenseNet121 model pretrained MIMIC-CXR dataset for deep learning-based multilabel classification using ground truth labels from radiology reports extracted using the CheXpert and CheXbert Labeler. We compared the performance of the 14 chest X-ray labels on the MIMIC-CXR and Veterans Healthcare Administration chest X-ray dataset (VA-CXR). The VA-CXR dataset comprises over 259k chest X-ray images spanning between the years 2010 and 2022. Results: The validation of ground truth and the assessment of multi-label classification performance across various NLP extraction tools revealed that the VA-CXR dataset exhibited lower disagreement rates than the MIMIC-CXR datasets. Additionally, there were notable differences in AUC scores between models utilizing CheXpert and CheXbert. When evaluating multi-label classification performance across different datasets, minimal domain shift was observed in unseen datasets, except for the label "Enlarged Cardiomediastinum." The study year's subgroup analyses exhibited the most significant variations in multi-label classification model performance. These findings underscore the importance of considering domain shifts in chest X-ray classification tasks, particularly concerning study years. Conclusion: Our study reveals the significant impact of domain shift and demographic factors on chest X-ray classification, emphasizing the need for improved transfer learning and equitable model development. Addressing these challenges is crucial for advancing medical imaging and enhancing patient care.
VISION: Toward a Standardized Process for Radiology Image Management at the National Level
Apr 29, 2024Abstract:The compilation and analysis of radiological images poses numerous challenges for researchers. The sheer volume of data as well as the computational needs of algorithms capable of operating on images are extensive. Additionally, the assembly of these images alone is difficult, as these exams may differ widely in terms of clinical context, structured annotation available for model training, modality, and patient identifiers. In this paper, we describe our experiences and challenges in establishing a trusted collection of radiology images linked to the United States Department of Veterans Affairs (VA) electronic health record database. We also discuss implications in making this repository research-ready for medical investigators. Key insights include uncovering the specific procedures required for transferring images from a clinical to a research-ready environment, as well as roadblocks and bottlenecks in this process that may hinder future efforts at automation.
LATTE: Label-efficient Incident Phenotyping from Longitudinal Electronic Health Records
May 19, 2023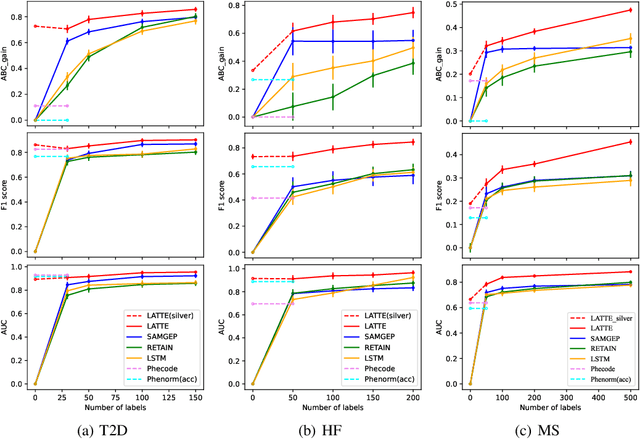
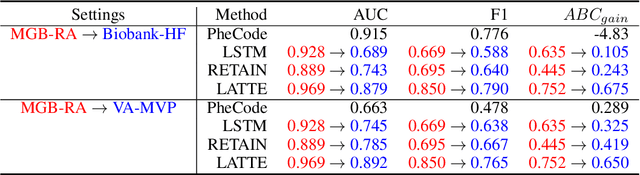
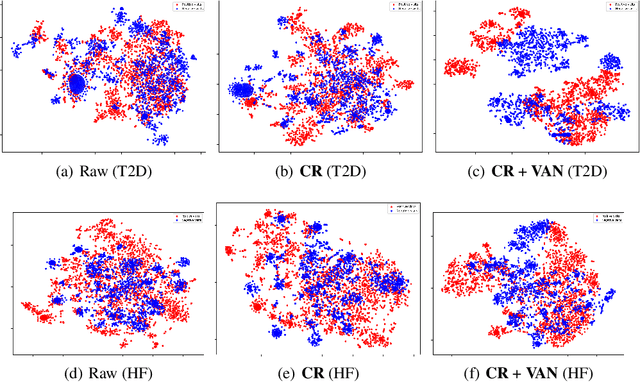
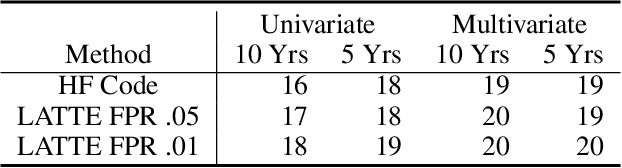
Abstract:Electronic health record (EHR) data are increasingly used to support real-world evidence (RWE) studies. Yet its ability to generate reliable RWE is limited by the lack of readily available precise information on the timing of clinical events such as the onset time of heart failure. We propose a LAbel-efficienT incidenT phEnotyping (LATTE) algorithm to accurately annotate the timing of clinical events from longitudinal EHR data. By leveraging the pre-trained semantic embedding vectors from large-scale EHR data as prior knowledge, LATTE selects predictive EHR features in a concept re-weighting module by mining their relationship to the target event and compresses their information into longitudinal visit embeddings through a visit attention learning network. LATTE employs a recurrent neural network to capture the sequential dependency between the target event and visit embeddings before/after it. To improve label efficiency, LATTE constructs highly informative longitudinal silver-standard labels from large-scale unlabeled patients to perform unsupervised pre-training and semi-supervised joint training. Finally, LATTE enhances cross-site portability via contrastive representation learning. LATTE is evaluated on three analyses: the onset of type-2 diabetes, heart failure, and the onset and relapses of multiple sclerosis. We use various evaluation metrics present in the literature including the $ABC_{gain}$, the proportion of reduction in the area between the observed event indicator and the predicted cumulative incidences in reference to the prediction per incident prevalence. LATTE consistently achieves substantial improvement over benchmark methods such as SAMGEP and RETAIN in all settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge