Xutao Guo
Building 3D In-Context Learning Universal Model in Neuroimaging
Mar 04, 2025Abstract:In-context learning (ICL), a type of universal model, demonstrates exceptional generalization across a wide range of tasks without retraining by leveraging task-specific guidance from context, making it particularly effective for the complex demands of neuroimaging. However, existing ICL models, which take 2D images as input, struggle to fully leverage the 3D anatomical structures in neuroimages, leading to a lack of global awareness and suboptimal performance. In this regard, we introduce Neuroverse3D, an ICL model capable of performing multiple neuroimaging tasks (e.g., segmentation, denoising, inpainting) in 3D. Neuroverse3D overcomes the large memory consumption due to 3D inputs through adaptive parallel-sequential context processing and a U-shape fusion strategy, allowing it to handle an unlimited number of context images. Additionally, we propose an optimized loss to balance multi-task training and enhance the focus on anatomical structures. Our study incorporates 43,674 3D scans from 19 neuroimaging datasets and evaluates Neuroverse3D on 14 diverse tasks using held-out test sets. The results demonstrate that Neuroverse3D significantly outperforms existing ICL models and closely matches the performance of task-specific models. The code and model weights are publicly released at: https://github.com/jiesihu/Neu3D.
Advancing Brain Imaging Analysis Step-by-step via Progressive Self-paced Learning
Jul 23, 2024



Abstract:Recent advancements in deep learning have shifted the development of brain imaging analysis. However, several challenges remain, such as heterogeneity, individual variations, and the contradiction between the high dimensionality and small size of brain imaging datasets. These issues complicate the learning process, preventing models from capturing intrinsic, meaningful patterns and potentially leading to suboptimal performance due to biases and overfitting. Curriculum learning (CL) presents a promising solution by organizing training examples from simple to complex, mimicking the human learning process, and potentially fostering the development of more robust and accurate models. Despite its potential, the inherent limitations posed by small initial training datasets present significant challenges, including overfitting and poor generalization. In this paper, we introduce the Progressive Self-Paced Distillation (PSPD) framework, employing an adaptive and progressive pacing and distillation mechanism. This allows for dynamic curriculum adjustments based on the states of both past and present models. The past model serves as a teacher, guiding the current model with gradually refined curriculum knowledge and helping prevent the loss of previously acquired knowledge. We validate PSPD's efficacy and adaptability across various convolutional neural networks using the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, underscoring its superiority in enhancing model performance and generalization capabilities. The source code for this approach will be released at https://github.com/Hrychen7/PSPD.
A Chebyshev Confidence Guided Source-Free Domain Adaptation Framework for Medical Image Segmentation
Oct 27, 2023



Abstract:Source-free domain adaptation (SFDA) aims to adapt models trained on a labeled source domain to an unlabeled target domain without the access to source data. In medical imaging scenarios, the practical significance of SFDA methods has been emphasized due to privacy concerns. Recent State-of-the-art SFDA methods primarily rely on self-training based on pseudo-labels (PLs). Unfortunately, PLs suffer from accuracy deterioration caused by domain shift, and thus limit the effectiveness of the adaptation process. To address this issue, we propose a Chebyshev confidence guided SFDA framework to accurately assess the reliability of PLs and generate self-improving PLs for self-training. The Chebyshev confidence is estimated by calculating probability lower bound of the PL confidence, given the prediction and the corresponding uncertainty. Leveraging the Chebyshev confidence, we introduce two confidence-guided denoising methods: direct denoising and prototypical denoising. Additionally, we propose a novel teacher-student joint training scheme (TJTS) that incorporates a confidence weighting module to improve PLs iteratively. The TJTS, in collaboration with the denoising methods, effectively prevents the propagation of noise and enhances the accuracy of PLs. Extensive experiments in diverse domain scenarios validate the effectiveness of our proposed framework and establish its superiority over state-of-the-art SFDA methods. Our paper contributes to the field of SFDA by providing a novel approach for precisely estimating the reliability of pseudo-labels and a framework for obtaining high-quality PLs, resulting in improved adaptation performance.
NexToU: Efficient Topology-Aware U-Net for Medical Image Segmentation
May 25, 2023Abstract:Convolutional neural networks (CNN) and Transformer variants have emerged as the leading medical image segmentation backbones. Nonetheless, due to their limitations in either preserving global image context or efficiently processing irregular shapes in visual objects, these backbones struggle to effectively integrate information from diverse anatomical regions and reduce inter-individual variability, particularly for the vasculature. Motivated by the successful breakthroughs of graph neural networks (GNN) in capturing topological properties and non-Euclidean relationships across various fields, we propose NexToU, a novel hybrid architecture for medical image segmentation. NexToU comprises improved Pool GNN and Swin GNN modules from Vision GNN (ViG) for learning both global and local topological representations while minimizing computational costs. To address the containment and exclusion relationships among various anatomical structures, we reformulate the topological interaction (TI) module based on the nature of binary trees, rapidly encoding the topological constraints into NexToU. Extensive experiments conducted on three datasets (including distinct imaging dimensions, disease types, and imaging modalities) demonstrate that our method consistently outperforms other state-of-the-art (SOTA) architectures. All the code is publicly available at https://github.com/PengchengShi1220/NexToU.
DECOR-NET: A COVID-19 Lung Infection Segmentation Network Improved by Emphasizing Low-level Features and Decorrelating Features
Feb 28, 2023Abstract:Since 2019, coronavirus Disease 2019 (COVID-19) has been widely spread and posed a serious threat to public health. Chest Computed Tomography (CT) holds great potential for screening and diagnosis of this disease. The segmentation of COVID-19 CT imaging can achieves quantitative evaluation of infections and tracks disease progression. COVID-19 infections are characterized by high heterogeneity and unclear boundaries, so capturing low-level features such as texture and intensity is critical for segmentation. However, segmentation networks that emphasize low-level features are still lacking. In this work, we propose a DECOR-Net capable of capturing more decorrelated low-level features. The channel re-weighting strategy is applied to obtain plenty of low-level features and the dependencies between channels are reduced by proposed decorrelation loss. Experiments show that DECOR-Net outperforms other cutting-edge methods and surpasses the baseline by 5.1% and 4.9% in terms of Dice coefficient and intersection over union. Moreover, the proposed decorrelation loss can improve the performance constantly under different settings. The Code is available at https://github.com/jiesihu/DECOR-Net.git.
Accelerating Diffusion Models via Pre-segmentation Diffusion Sampling for Medical Image Segmentation
Oct 27, 2022



Abstract:Based on the Denoising Diffusion Probabilistic Model (DDPM), medical image segmentation can be described as a conditional image generation task, which allows to compute pixel-wise uncertainty maps of the segmentation and allows an implicit ensemble of segmentations to boost the segmentation performance. However, DDPM requires many iterative denoising steps to generate segmentations from Gaussian noise, resulting in extremely inefficient inference. To mitigate the issue, we propose a principled acceleration strategy, called pre-segmentation diffusion sampling DDPM (PD-DDPM), which is specially used for medical image segmentation. The key idea is to obtain pre-segmentation results based on a separately trained segmentation network, and construct noise predictions (non-Gaussian distribution) according to the forward diffusion rule. We can then start with noisy predictions and use fewer reverse steps to generate segmentation results. Experiments show that PD-DDPM yields better segmentation results over representative baseline methods even if the number of reverse steps is significantly reduced. Moreover, PD-DDPM is orthogonal to existing advanced segmentation models, which can be combined to further improve the segmentation performance.
Multi-modal Dynamic Graph Network: Coupling Structural and Functional Connectome for Disease Diagnosis and Classification
Oct 25, 2022Abstract:Multi-modal neuroimaging technology has greatlly facilitated the efficiency and diagnosis accuracy, which provides complementary information in discovering objective disease biomarkers. Conventional deep learning methods, e.g. convolutional neural networks, overlook relationships between nodes and fail to capture topological properties in graphs. Graph neural networks have been proven to be of great importance in modeling brain connectome networks and relating disease-specific patterns. However, most existing graph methods explicitly require known graph structures, which are not available in the sophisticated brain system. Especially in heterogeneous multi-modal brain networks, there exists a great challenge to model interactions among brain regions in consideration of inter-modal dependencies. In this study, we propose a Multi-modal Dynamic Graph Convolution Network (MDGCN) for structural and functional brain network learning. Our method benefits from modeling inter-modal representations and relating attentive multi-model associations into dynamic graphs with a compositional correspondence matrix. Moreover, a bilateral graph convolution layer is proposed to aggregate multi-modal representations in terms of multi-modal associations. Extensive experiments on three datasets demonstrate the superiority of our proposed method in terms of disease classification, with the accuracy of 90.4%, 85.9% and 98.3% in predicting Mild Cognitive Impairment (MCI), Parkinson's disease (PD), and schizophrenia (SCHZ) respectively. Furthermore, our statistical evaluations on the correspondence matrix exhibit a high correspondence with previous evidence of biomarkers.
Estimating Brain Age with Global and Local Dependencies
Sep 19, 2022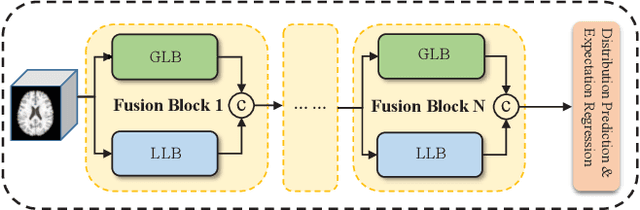
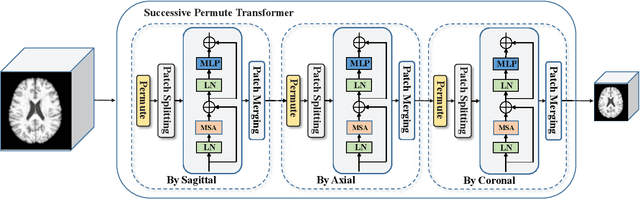
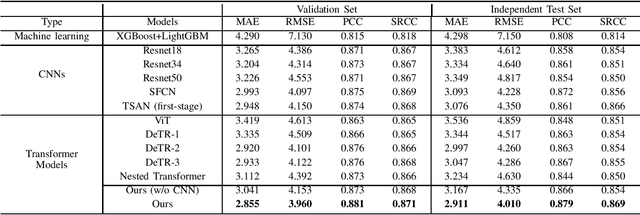
Abstract:The brain age has been proven to be a phenotype of relevance to cognitive performance and brain disease. Achieving accurate brain age prediction is an essential prerequisite for optimizing the predicted brain-age difference as a biomarker. As a comprehensive biological characteristic, the brain age is hard to be exploited accurately with models using feature engineering and local processing such as local convolution and recurrent operations that process one local neighborhood at a time. Instead, Vision Transformers learn global attentive interaction of patch tokens, introducing less inductive bias and modeling long-range dependencies. In terms of this, we proposed a novel network for learning brain age interpreting with global and local dependencies, where the corresponding representations are captured by Successive Permuted Transformer (SPT) and convolution blocks. The SPT brings computation efficiency and locates the 3D spatial information indirectly via continuously encoding 2D slices from different views. Finally, we collect a large cohort of 22645 subjects with ages ranging from 14 to 97 and our network performed the best among a series of deep learning methods, yielding a mean absolute error (MAE) of 2.855 in validation set, and 2.911 in an independent test set.
Uncertainty Quantification in Medical Image Segmentation with Multi-decoder U-Net
Sep 15, 2021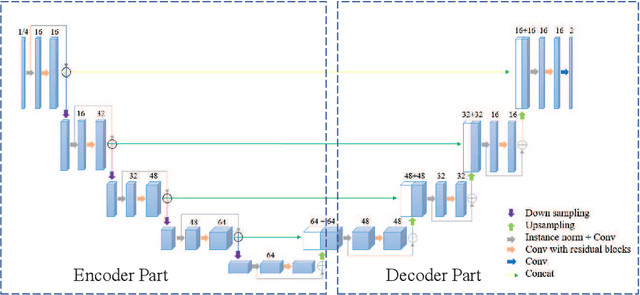
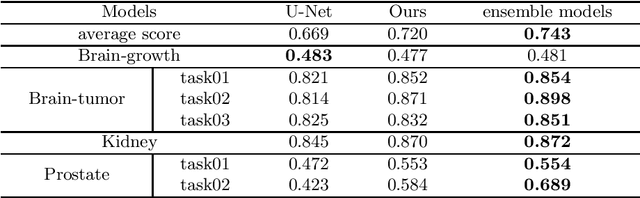
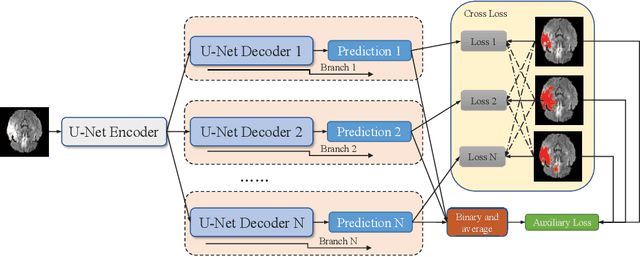
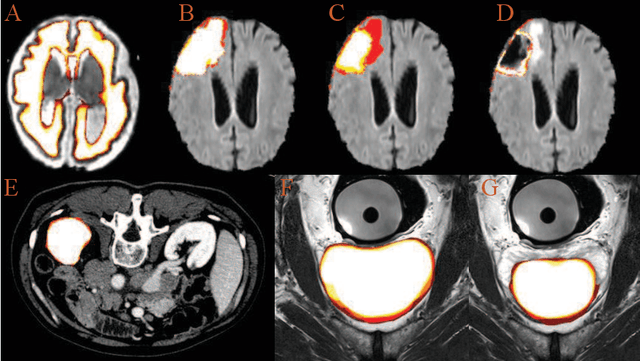
Abstract:Accurate medical image segmentation is crucial for diagnosis and analysis. However, the models without calibrated uncertainty estimates might lead to errors in downstream analysis and exhibit low levels of robustness. Estimating the uncertainty in the measurement is vital to making definite, informed conclusions. Especially, it is difficult to make accurate predictions on ambiguous areas and focus boundaries for both models and radiologists, even harder to reach a consensus with multiple annotations. In this work, the uncertainty under these areas is studied, which introduces significant information with anatomical structure and is as important as segmentation performance. We exploit the medical image segmentation uncertainty quantification by measuring segmentation performance with multiple annotations in a supervised learning manner and propose a U-Net based architecture with multiple decoders, where the image representation is encoded with the same encoder, and segmentation referring to each annotation is estimated with multiple decoders. Nevertheless, a cross-loss function is proposed for bridging the gap between different branches. The proposed architecture is trained in an end-to-end manner and able to improve predictive uncertainty estimates. The model achieves comparable performance with fewer parameters to the integrated training model that ranked the runner-up in the MICCAI-QUBIQ 2020 challenge.
SeFM: A Sequential Feature Point Matching Algorithm for Object 3D Reconstruction
Dec 07, 2018
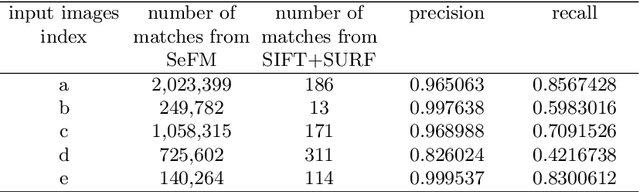
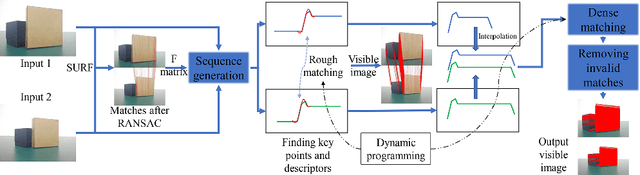

Abstract:3D reconstruction is a fundamental issue in many applications and the feature point matching problem is a key step while reconstructing target objects. Conventional algorithms can only find a small number of feature points from two images which is quite insufficient for reconstruction. To overcome this problem, we propose SeFM a sequential feature point matching algorithm. We first utilize the epipolar geometry to find the epipole of each image. Rotating along the epipole, we generate a set of the epipolar lines and reserve those intersecting with the input image. Next, a rough matching phase, followed by a dense matching phase, is applied to find the matching dot-pairs using dynamic programming. Furthermore, we also remove wrong matching dot-pairs by calculating the validity. Experimental results illustrate that SeFM can achieve around 1,000 to 10,000 times matching dot-pairs, depending on individual image, compared to conventional algorithms and the object reconstruction with only two images is semantically visible. Moreover, it outperforms conventional algorithms, such as SIFT and SURF, regarding precision and recall.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge