Xinghua Lu
Knowledge-guided Contextual Gene Set Analysis Using Large Language Models
Jun 04, 2025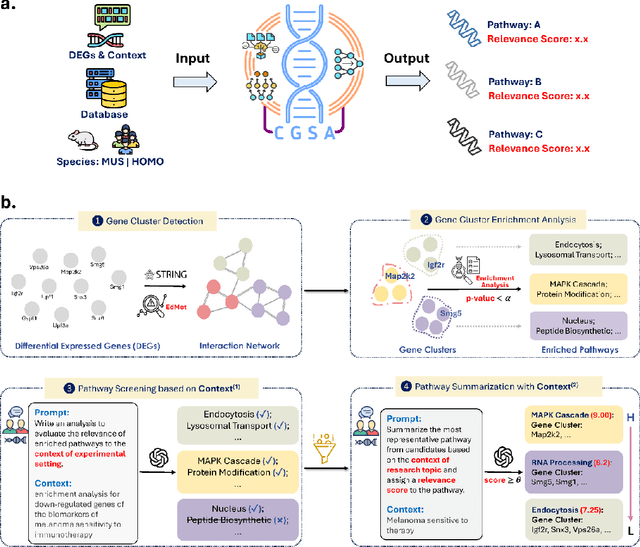

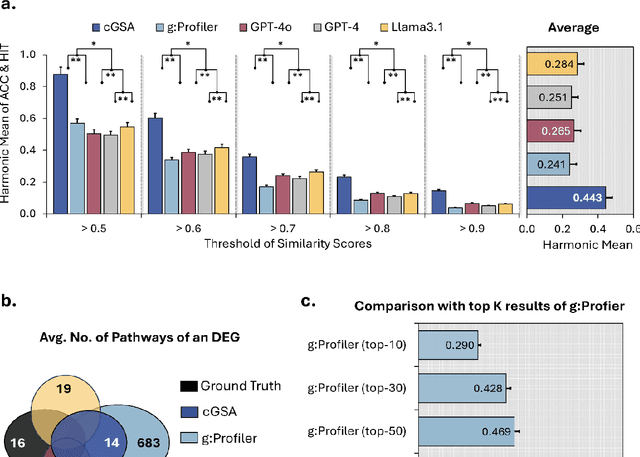

Abstract:Gene set analysis (GSA) is a foundational approach for interpreting genomic data of diseases by linking genes to biological processes. However, conventional GSA methods overlook clinical context of the analyses, often generating long lists of enriched pathways with redundant, nonspecific, or irrelevant results. Interpreting these requires extensive, ad-hoc manual effort, reducing both reliability and reproducibility. To address this limitation, we introduce cGSA, a novel AI-driven framework that enhances GSA by incorporating context-aware pathway prioritization. cGSA integrates gene cluster detection, enrichment analysis, and large language models to identify pathways that are not only statistically significant but also biologically meaningful. Benchmarking on 102 manually curated gene sets across 19 diseases and ten disease-related biological mechanisms shows that cGSA outperforms baseline methods by over 30%, with expert validation confirming its increased precision and interpretability. Two independent case studies in melanoma and breast cancer further demonstrate its potential to uncover context-specific insights and support targeted hypothesis generation.
Understanding Heart-Failure Patients EHR Clinical Features via SHAP Interpretation of Tree-Based Machine Learning Model Predictions
Mar 20, 2021


Abstract:Heart failure (HF) is a major cause of mortality. Accurately monitoring HF progress and adjust therapies are critical for improving patient outcomes. An experienced cardiologist can make accurate HF stage diagnoses based on combination of symptoms, signs, and lab results from the electronic health records (EHR) of a patient, without directly measuring heart function. We examined whether machine learning models, more specifically the XGBoost model, can accurately predict patient stage based on EHR, and we further applied the SHapley Additive exPlanations (SHAP) framework to identify informative features and their interpretations. Our results indicate that based on structured data from EHR, our models could predict patients' ejection fraction (EF) scores with moderate accuracy. SHAP analyses identified informative features and revealed potential clinical subtypes of HF. Our findings provide insights on how to design computing systems to accurately monitor disease progression of HF patients through continuously mining patients' EHR data.
Learning Latent Causal Structures with a Redundant Input Neural Network
Mar 29, 2020
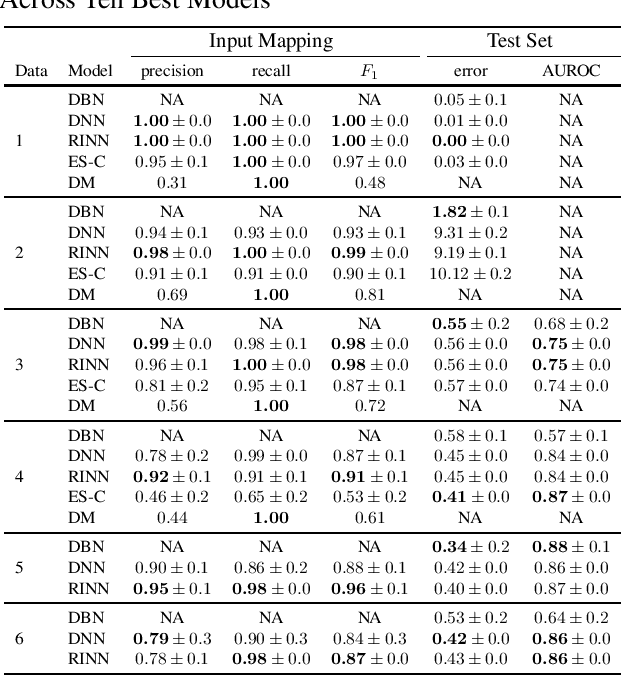


Abstract:Most causal discovery algorithms find causal structure among a set of observed variables. Learning the causal structure among latent variables remains an important open problem, particularly when using high-dimensional data. In this paper, we address a problem for which it is known that inputs cause outputs, and these causal relationships are encoded by a causal network among a set of an unknown number of latent variables. We developed a deep learning model, which we call a redundant input neural network (RINN), with a modified architecture and a regularized objective function to find causal relationships between input, hidden, and output variables. More specifically, our model allows input variables to directly interact with all latent variables in a neural network to influence what information the latent variables should encode in order to generate the output variables accurately. In this setting, the direct connections between input and latent variables makes the latent variables partially interpretable; furthermore, the connectivity among the latent variables in the neural network serves to model their potential causal relationships to each other and to the output variables. A series of simulation experiments provide support that the RINN method can successfully recover latent causal structure between input and output variables.
Supervised Vector Quantized Variational Autoencoder for Learning Interpretable Global Representations
Sep 29, 2019
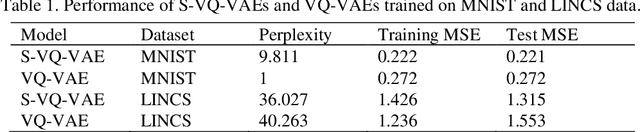

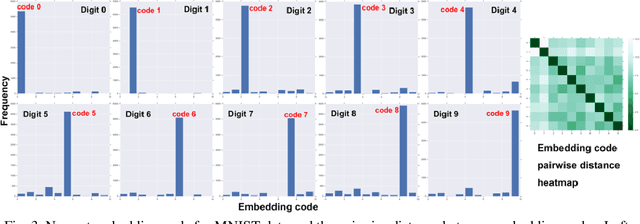
Abstract:Learning interpretable representations of data remains a central challenge in deep learning. When training a deep generative model, the observed data are often associated with certain categorical labels, and, in parallel with learning to regenerate data and simulate new data, learning an interpretable representation of each class of data is also a process of acquiring knowledge. Here, we present a novel generative model, referred to as the Supervised Vector Quantized Variational AutoEncoder (S-VQ-VAE), which combines the power of supervised and unsupervised learning to obtain a unique, interpretable global representation for each class of data. Compared with conventional generative models, our model has three key advantages: first, it is an integrative model that can simultaneously learn a feature representation for individual data point and a global representation for each class of data; second, the learning of global representations with embedding codes is guided by supervised information, which clearly defines the interpretation of each code; and third, the global representations capture crucial characteristics of different classes, which reveal similarity and differences of statistical structures underlying different groups of data. We evaluated the utility of S-VQ-VAE on a machine learning benchmark dataset, the MNIST dataset, and on gene expression data from the Library of Integrated Network-Based Cellular Signatures (LINCS). We proved that S-VQ-VAE was able to learn the global genetic characteristics of samples perturbed by the same class of perturbagen (PCL), and further revealed the mechanism correlations between PCLs. Such knowledge is crucial for promoting new drug development for complex diseases like cancer.
PubMedQA: A Dataset for Biomedical Research Question Answering
Sep 13, 2019


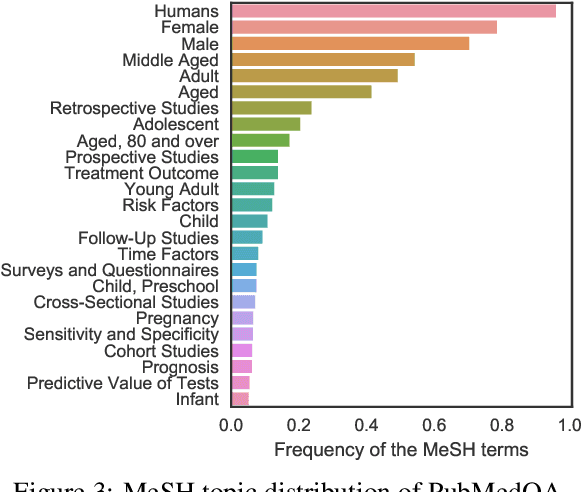
Abstract:We introduce PubMedQA, a novel biomedical question answering (QA) dataset collected from PubMed abstracts. The task of PubMedQA is to answer research questions with yes/no/maybe (e.g.: Do preoperative statins reduce atrial fibrillation after coronary artery bypass grafting?) using the corresponding abstracts. PubMedQA has 1k expert-annotated, 61.2k unlabeled and 211.3k artificially generated QA instances. Each PubMedQA instance is composed of (1) a question which is either an existing research article title or derived from one, (2) a context which is the corresponding abstract without its conclusion, (3) a long answer, which is the conclusion of the abstract and, presumably, answers the research question, and (4) a yes/no/maybe answer which summarizes the conclusion. PubMedQA is the first QA dataset where reasoning over biomedical research texts, especially their quantitative contents, is required to answer the questions. Our best performing model, multi-phase fine-tuning of BioBERT with long answer bag-of-word statistics as additional supervision, achieves 68.1% accuracy, compared to single human performance of 78.0% accuracy and majority-baseline of 55.2% accuracy, leaving much room for improvement. PubMedQA is publicly available at https://pubmedqa.github.io.
Deep Contextualized Biomedical Abbreviation Expansion
Jun 08, 2019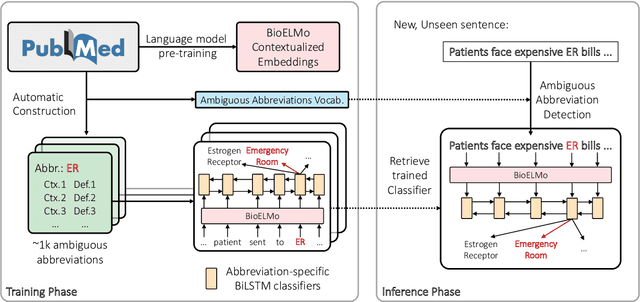

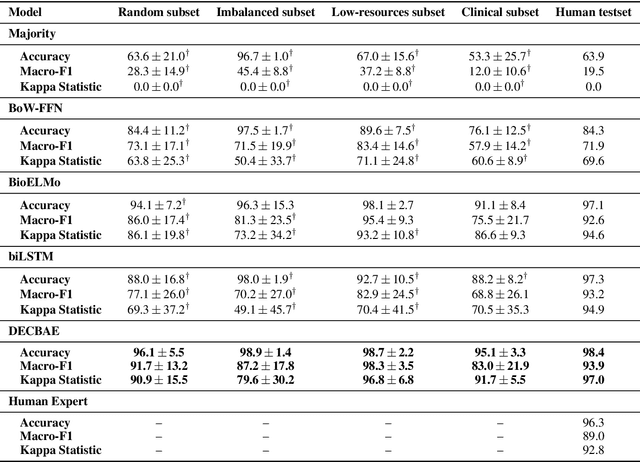
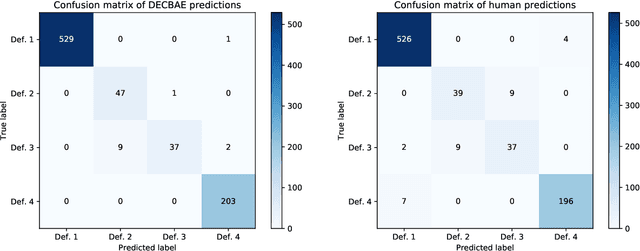
Abstract:Automatic identification and expansion of ambiguous abbreviations are essential for biomedical natural language processing applications, such as information retrieval and question answering systems. In this paper, we present DEep Contextualized Biomedical. Abbreviation Expansion (DECBAE) model. DECBAE automatically collects substantial and relatively clean annotated contexts for 950 ambiguous abbreviations from PubMed abstracts using a simple heuristic. Then it utilizes BioELMo to extract the contextualized features of words, and feed those features to abbreviation-specific bidirectional LSTMs, where the hidden states of the ambiguous abbreviations are used to assign the exact definitions. Our DECBAE model outperforms other baselines by large margins, achieving average accuracy of 0.961 and macro-F1 of 0.917 on the dataset. It also surpasses human performance for expanding a sample abbreviation, and remains robust in imbalanced, low-resources and clinical settings.
Probing Biomedical Embeddings from Language Models
Apr 03, 2019

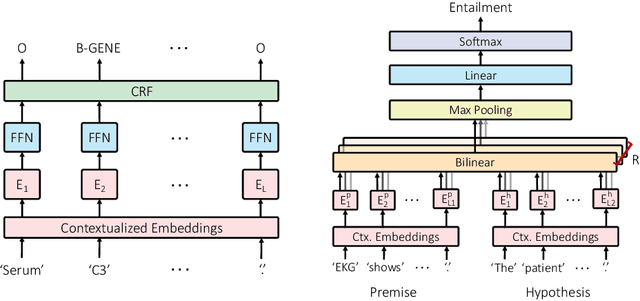

Abstract:Contextualized word embeddings derived from pre-trained language models (LMs) show significant improvements on downstream NLP tasks. Pre-training on domain-specific corpora, such as biomedical articles, further improves their performance. In this paper, we conduct probing experiments to determine what additional information is carried intrinsically by the in-domain trained contextualized embeddings. For this we use the pre-trained LMs as fixed feature extractors and restrict the downstream task models to not have additional sequence modeling layers. We compare BERT, ELMo, BioBERT and BioELMo, a biomedical version of ELMo trained on 10M PubMed abstracts. Surprisingly, while fine-tuned BioBERT is better than BioELMo in biomedical NER and NLI tasks, as a fixed feature extractor BioELMo outperforms BioBERT in our probing tasks. We use visualization and nearest neighbor analysis to show that better encoding of entity-type and relational information leads to this superiority.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge