Xianqi Li
Missing Data Estimation for MR Spectroscopic Imaging via Mask-Free Deep Learning Methods
May 11, 2025Abstract:Magnetic Resonance Spectroscopic Imaging (MRSI) is a powerful tool for non-invasive mapping of brain metabolites, providing critical insights into neurological conditions. However, its utility is often limited by missing or corrupted data due to motion artifacts, magnetic field inhomogeneities, or failed spectral fitting-especially in high resolution 3D acquisitions. To address this, we propose the first deep learning-based, mask-free framework for estimating missing data in MRSI metabolic maps. Unlike conventional restoration methods that rely on explicit masks to identify missing regions, our approach implicitly detects and estimates these areas using contextual spatial features through 2D and 3D U-Net architectures. We also introduce a progressive training strategy to enhance robustness under varying levels of data degradation. Our method is evaluated on both simulated and real patient datasets and consistently outperforms traditional interpolation techniques such as cubic and linear interpolation. The 2D model achieves an MSE of 0.002 and an SSIM of 0.97 with 20% missing voxels, while the 3D model reaches an MSE of 0.001 and an SSIM of 0.98 with 15% missing voxels. Qualitative results show improved fidelity in estimating missing data, particularly in metabolically heterogeneous regions and ventricular regions. Importantly, our model generalizes well to real-world datasets without requiring retraining or mask input. These findings demonstrate the effectiveness and broad applicability of mask-free deep learning for MRSI restoration, with strong potential for clinical and research integration.
A Low-complexity Structured Neural Network to Realize States of Dynamical Systems
Mar 31, 2025Abstract:Data-driven learning is rapidly evolving and places a new perspective on realizing state-space dynamical systems. However, dynamical systems derived from nonlinear ordinary differential equations (ODEs) suffer from limitations in computational efficiency. Thus, this paper stems from data-driven learning to advance states of dynamical systems utilizing a structured neural network (StNN). The proposed learning technique also seeks to identify an optimal, low-complexity operator to solve dynamical systems, the so-called Hankel operator, derived from time-delay measurements. Thus, we utilize the StNN based on the Hankel operator to solve dynamical systems as an alternative to existing data-driven techniques. We show that the proposed StNN reduces the number of parameters and computational complexity compared with the conventional neural networks and also with the classical data-driven techniques, such as Sparse Identification of Nonlinear Dynamics (SINDy) and Hankel Alternative view of Koopman (HAVOK), which is commonly known as delay-Dynamic Mode Decomposition(DMD) or Hankel-DMD. More specifically, we present numerical simulations to solve dynamical systems utilizing the StNN based on the Hankel operator beginning from the fundamental Lotka-Volterra model, where we compare the StNN with the LEarning Across Dynamical Systems (LEADS), and extend our analysis to highly nonlinear and chaotic Lorenz systems, comparing the StNN with conventional neural networks, SINDy, and HAVOK. Hence, we show that the proposed StNN paves the way for realizing state-space dynamical systems with a low-complexity learning algorithm, enabling prediction and understanding of future states.
A Low-complexity Structured Neural Network Approach to Intelligently Realize Wideband Multi-beam Beamformers
Mar 26, 2025Abstract:True-time-delay (TTD) beamformers can produce wideband, squint-free beams in both analog and digital signal domains, unlike frequency-dependent FFT beams. Our previous work showed that TTD beamformers can be efficiently realized using the elements of delay Vandermonde matrix (DVM), answering the longstanding beam-squint problem. Thus, building on our work on classical algorithms based on DVM, we propose neural network (NN) architecture to realize wideband multi-beam beamformers using structure-imposed weight matrices and submatrices. The structure and sparsity of the weight matrices and submatrices are shown to reduce the space and computational complexities of the NN greatly. The proposed network architecture has O(pLM logM) complexity compared to a conventional fully connected L-layers network with O(M2L) complexity, where M is the number of nodes in each layer of the network, p is the number of submatrices per layer, and M >> p. We will show numerical simulations in the 24 GHz to 32 GHz range to demonstrate the numerical feasibility of realizing wideband multi-beam beamformers using the proposed neural architecture. We also show the complexity reduction of the proposed NN and compare that with fully connected NNs, to show the efficiency of the proposed architecture without sacrificing accuracy. The accuracy of the proposed NN architecture was shown using the mean squared error, which is based on an objective function of the weight matrices and beamformed signals of antenna arrays, while also normalizing nodes. The proposed NN architecture shows a low-complexity NN realizing wideband multi-beam beamformers in real-time for low-complexity intelligent systems.
SeUNet-Trans: A Simple yet Effective UNet-Transformer Model for Medical Image Segmentation
Oct 22, 2023Abstract:Automated medical image segmentation is becoming increasingly crucial in modern clinical practice, driven by the growing demand for precise diagnoses, the push towards personalized treatment plans, and advancements in machine learning algorithms, especially the incorporation of deep learning methods. While convolutional neural networks (CNNs) have been prevalent among these methods, the remarkable potential of Transformer-based models for computer vision tasks is gaining more acknowledgment. To harness the advantages of both CNN-based and Transformer-based models, we propose a simple yet effective UNet-Transformer (seUNet-Trans) model for medical image segmentation. In our approach, the UNet model is designed as a feature extractor to generate multiple feature maps from the input images, and these maps are propagated into a bridge layer, which sequentially connects the UNet and the Transformer. In this stage, we employ the pixel-level embedding technique without position embedding vectors to make the model more efficient. Moreover, we applied spatial-reduction attention in the Transformer to reduce the computational/memory overhead. By leveraging the UNet architecture and the self-attention mechanism, our model not only preserves both local and global context information but also captures long-range dependencies between input elements. The proposed model is extensively experimented on five medical image segmentation datasets, including polyp segmentation, to demonstrate its efficacy. A comparison with several state-of-the-art segmentation models on these datasets shows the superior performance of seUNet-Trans.
A Comprehensive Review of Generative AI in Healthcare
Oct 01, 2023



Abstract:The advancement of Artificial Intelligence (AI) has catalyzed revolutionary changes across various sectors, notably in healthcare. Among the significant developments in this field are the applications of generative AI models, specifically transformers and diffusion models. These models have played a crucial role in analyzing diverse forms of data, including medical imaging (encompassing image reconstruction, image-to-image translation, image generation, and image classification), protein structure prediction, clinical documentation, diagnostic assistance, radiology interpretation, clinical decision support, medical coding, and billing, as well as drug design and molecular representation. Such applications have enhanced clinical diagnosis, data reconstruction, and drug synthesis. This review paper aims to offer a thorough overview of the generative AI applications in healthcare, focusing on transformers and diffusion models. Additionally, we propose potential directions for future research to tackle the existing limitations and meet the evolving demands of the healthcare sector. Intended to serve as a comprehensive guide for researchers and practitioners interested in the healthcare applications of generative AI, this review provides valuable insights into the current state of the art, challenges faced, and prospective future directions.
Adaptive Reorganization of Neural Pathways for Continual Learning with Hybrid Spiking Neural Networks
Sep 18, 2023



Abstract:The human brain can self-organize rich and diverse sparse neural pathways to incrementally master hundreds of cognitive tasks. However, most existing continual learning algorithms for deep artificial and spiking neural networks are unable to adequately auto-regulate the limited resources in the network, which leads to performance drop along with energy consumption rise as the increase of tasks. In this paper, we propose a brain-inspired continual learning algorithm with adaptive reorganization of neural pathways, which employs Self-Organizing Regulation networks to reorganize the single and limited Spiking Neural Network (SOR-SNN) into rich sparse neural pathways to efficiently cope with incremental tasks. The proposed model demonstrates consistent superiority in performance, energy consumption, and memory capacity on diverse continual learning tasks ranging from child-like simple to complex tasks, as well as on generalized CIFAR100 and ImageNet datasets. In particular, the SOR-SNN model excels at learning more complex tasks as well as more tasks, and is able to integrate the past learned knowledge with the information from the current task, showing the backward transfer ability to facilitate the old tasks. Meanwhile, the proposed model exhibits self-repairing ability to irreversible damage and for pruned networks, could automatically allocate new pathway from the retained network to recover memory for forgotten knowledge.
Deep Learning-based Prediction of Stress and Strain Maps in Arterial Walls for Improved Cardiovascular Risk Assessment
Aug 03, 2023



Abstract:This study investigated the potential of end-to-end deep learning tools as a more effective substitute for FEM in predicting stress-strain fields within 2D cross sections of arterial wall. We first proposed a U-Net based fully convolutional neural network (CNN) to predict the von Mises stress and strain distribution based on the spatial arrangement of calcification within arterial wall cross-sections. Further, we developed a conditional generative adversarial network (cGAN) to enhance, particularly from the perceptual perspective, the prediction accuracy of stress and strain field maps for arterial walls with various calcification quantities and spatial configurations. On top of U-Net and cGAN, we also proposed their ensemble approaches, respectively, to further improve the prediction accuracy of field maps. Our dataset, consisting of input and output images, was generated by implementing boundary conditions and extracting stress-strain field maps. The trained U-Net models can accurately predict von Mises stress and strain fields, with structural similarity index scores (SSIM) of 0.854 and 0.830 and mean squared errors of 0.017 and 0.018 for stress and strain, respectively, on a reserved test set. Meanwhile, the cGAN models in a combination of ensemble and transfer learning techniques demonstrate high accuracy in predicting von Mises stress and strain fields, as evidenced by SSIM scores of 0.890 for stress and 0.803 for strain. Additionally, mean squared errors of 0.008 for stress and 0.017 for strain further support the model's performance on a designated test set. Overall, this study developed a surrogate model for finite element analysis, which can accurately and efficiently predict stress-strain fields of arterial walls regardless of complex geometries and boundary conditions.
Breast Tumor Classification Based on Decision Information Genes and Inverse Projection Sparse Representation
Apr 17, 2018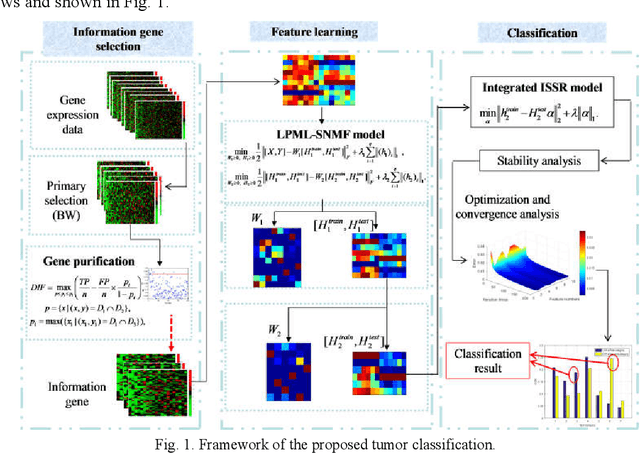
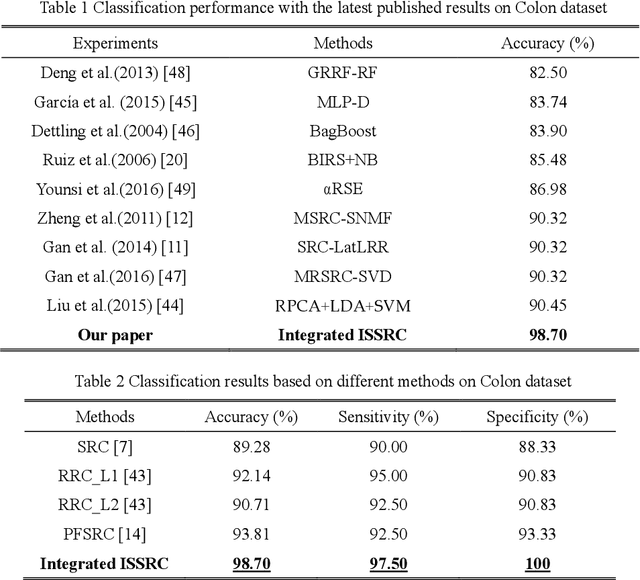
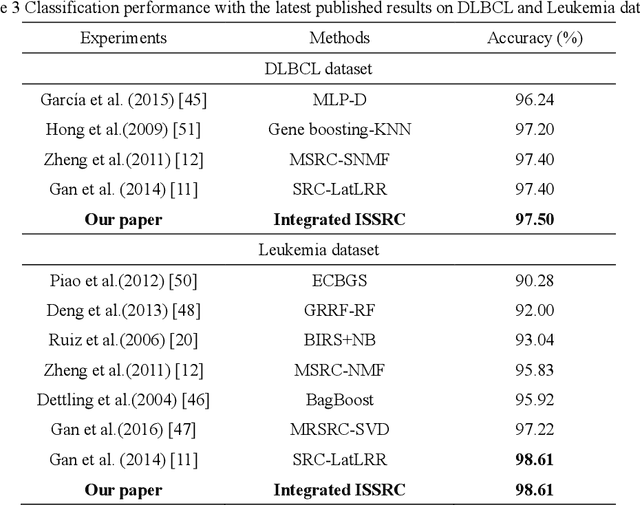
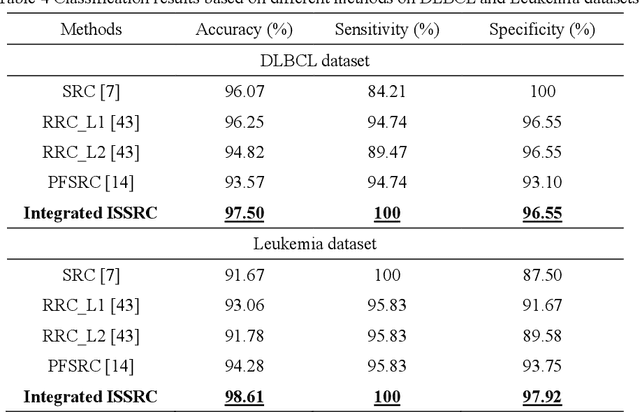
Abstract:Microarray gene expression data-based breast tumor classification is an active and challenging issue. In this paper, a robust framework of breast tumor recognition is presented aiming at reducing clinical misdiagnosis rate and exploiting available information in existing samples. A wrapper gene selection method is established from a new perspective of reducing clinical misdiagnosis rate. The further feature selection of information genes is achieved using the modified NMF model, which is rooted in the use of hierarchical learning and layer-wise pre-training strategy in deep learning. For completing the classification, an inverse projection sparse representation (IPSR) model is constructed to exploit information embedded in existing samples, especially in the test ones. Moreover, the IPSR model is optimized through generalized ADMM and the corresponding convergence is analyzed. Extensive experiments on public microarray gene expression datasets show that the proposed method is stable and effective for breast tumor classification. Compared to the latest open literature, there is 14% higher in classification accuracy. Specificity and sensitivity achieve 94.17% and 97.5%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge