Wei Long
From Local Windows to Adaptive Candidates via Individualized Exploratory: Rethinking Attention for Image Super-Resolution
Jan 13, 2026Abstract:Single Image Super-Resolution (SISR) is a fundamental computer vision task that aims to reconstruct a high-resolution (HR) image from a low-resolution (LR) input. Transformer-based methods have achieved remarkable performance by modeling long-range dependencies in degraded images. However, their feature-intensive attention computation incurs high computational cost. To improve efficiency, most existing approaches partition images into fixed groups and restrict attention within each group. Such group-wise attention overlooks the inherent asymmetry in token similarities, thereby failing to enable flexible and token-adaptive attention computation. To address this limitation, we propose the Individualized Exploratory Transformer (IET), which introduces a novel Individualized Exploratory Attention (IEA) mechanism that allows each token to adaptively select its own content-aware and independent attention candidates. This token-adaptive and asymmetric design enables more precise information aggregation while maintaining computational efficiency. Extensive experiments on standard SR benchmarks demonstrate that IET achieves state-of-the-art performance under comparable computational complexity.
IDESplat: Iterative Depth Probability Estimation for Generalizable 3D Gaussian Splatting
Jan 07, 2026Abstract:Generalizable 3D Gaussian Splatting aims to directly predict Gaussian parameters using a feed-forward network for scene reconstruction. Among these parameters, Gaussian means are particularly difficult to predict, so depth is usually estimated first and then unprojected to obtain the Gaussian sphere centers. Existing methods typically rely solely on a single warp to estimate depth probability, which hinders their ability to fully leverage cross-view geometric cues, resulting in unstable and coarse depth maps. To address this limitation, we propose IDESplat, which iteratively applies warp operations to boost depth probability estimation for accurate Gaussian mean prediction. First, to eliminate the inherent instability of a single warp, we introduce a Depth Probability Boosting Unit (DPBU) that integrates epipolar attention maps produced by cascading warp operations in a multiplicative manner. Next, we construct an iterative depth estimation process by stacking multiple DPBUs, progressively identifying potential depth candidates with high likelihood. As IDESplat iteratively boosts depth probability estimates and updates the depth candidates, the depth map is gradually refined, resulting in accurate Gaussian means. We conduct experiments on RealEstate10K, ACID, and DL3DV. IDESplat achieves outstanding reconstruction quality and state-of-the-art performance with real-time efficiency. On RE10K, it outperforms DepthSplat by 0.33 dB in PSNR, using only 10.7% of the parameters and 70% of the memory. Additionally, our IDESplat improves PSNR by 2.95 dB over DepthSplat on the DTU dataset in cross-dataset experiments, demonstrating its strong generalization ability.
MVAR: Visual Autoregressive Modeling with Scale and Spatial Markovian Conditioning
May 19, 2025Abstract:Essential to visual generation is efficient modeling of visual data priors. Conventional next-token prediction methods define the process as learning the conditional probability distribution of successive tokens. Recently, next-scale prediction methods redefine the process to learn the distribution over multi-scale representations, significantly reducing generation latency. However, these methods condition each scale on all previous scales and require each token to consider all preceding tokens, exhibiting scale and spatial redundancy. To better model the distribution by mitigating redundancy, we propose Markovian Visual AutoRegressive modeling (MVAR), a novel autoregressive framework that introduces scale and spatial Markov assumptions to reduce the complexity of conditional probability modeling. Specifically, we introduce a scale-Markov trajectory that only takes as input the features of adjacent preceding scale for next-scale prediction, enabling the adoption of a parallel training strategy that significantly reduces GPU memory consumption. Furthermore, we propose spatial-Markov attention, which restricts the attention of each token to a localized neighborhood of size k at corresponding positions on adjacent scales, rather than attending to every token across these scales, for the pursuit of reduced modeling complexity. Building on these improvements, we reduce the computational complexity of attention calculation from O(N^2) to O(Nk), enabling training with just eight NVIDIA RTX 4090 GPUs and eliminating the need for KV cache during inference. Extensive experiments on ImageNet demonstrate that MVAR achieves comparable or superior performance with both small model trained from scratch and large fine-tuned models, while reducing the average GPU memory footprint by 3.0x.
Small Clips, Big Gains: Learning Long-Range Refocused Temporal Information for Video Super-Resolution
May 04, 2025Abstract:Video super-resolution (VSR) can achieve better performance compared to single image super-resolution by additionally leveraging temporal information. In particular, the recurrent-based VSR model exploits long-range temporal information during inference and achieves superior detail restoration. However, effectively learning these long-term dependencies within long videos remains a key challenge. To address this, we propose LRTI-VSR, a novel training framework for recurrent VSR that efficiently leverages Long-Range Refocused Temporal Information. Our framework includes a generic training strategy that utilizes temporal propagation features from long video clips while training on shorter video clips. Additionally, we introduce a refocused intra&inter-frame transformer block which allows the VSR model to selectively prioritize useful temporal information through its attention module while further improving inter-frame information utilization in the FFN module. We evaluate LRTI-VSR on both CNN and transformer-based VSR architectures, conducting extensive ablation studies to validate the contribution of each component. Experiments on long-video test sets demonstrate that LRTI-VSR achieves state-of-the-art performance while maintaining training and computational efficiency.
Progressive Focused Transformer for Single Image Super-Resolution
Mar 26, 2025Abstract:Transformer-based methods have achieved remarkable results in image super-resolution tasks because they can capture non-local dependencies in low-quality input images. However, this feature-intensive modeling approach is computationally expensive because it calculates the similarities between numerous features that are irrelevant to the query features when obtaining attention weights. These unnecessary similarity calculations not only degrade the reconstruction performance but also introduce significant computational overhead. How to accurately identify the features that are important to the current query features and avoid similarity calculations between irrelevant features remains an urgent problem. To address this issue, we propose a novel and effective Progressive Focused Transformer (PFT) that links all isolated attention maps in the network through Progressive Focused Attention (PFA) to focus attention on the most important tokens. PFA not only enables the network to capture more critical similar features, but also significantly reduces the computational cost of the overall network by filtering out irrelevant features before calculating similarities. Extensive experiments demonstrate the effectiveness of the proposed method, achieving state-of-the-art performance on various single image super-resolution benchmarks.
Threshold Attention Network for Semantic Segmentation of Remote Sensing Images
Jan 14, 2025



Abstract:Semantic segmentation of remote sensing images is essential for various applications, including vegetation monitoring, disaster management, and urban planning. Previous studies have demonstrated that the self-attention mechanism (SA) is an effective approach for designing segmentation networks that can capture long-range pixel dependencies. SA enables the network to model the global dependencies between the input features, resulting in improved segmentation outcomes. However, the high density of attentional feature maps used in this mechanism causes exponential increases in computational complexity. Additionally, it introduces redundant information that negatively impacts the feature representation. Inspired by traditional threshold segmentation algorithms, we propose a novel threshold attention mechanism (TAM). This mechanism significantly reduces computational effort while also better modeling the correlation between different regions of the feature map. Based on TAM, we present a threshold attention network (TANet) for semantic segmentation. TANet consists of an attentional feature enhancement module (AFEM) for global feature enhancement of shallow features and a threshold attention pyramid pooling module (TAPP) for acquiring feature information at different scales for deep features. We have conducted extensive experiments on the ISPRS Vaihingen and Potsdam datasets. The results demonstrate the validity and superiority of our proposed TANet compared to the most state-of-the-art models.
MoCha-Stereo: Motif Channel Attention Network for Stereo Matching
Apr 11, 2024Abstract:Learning-based stereo matching techniques have made significant progress. However, existing methods inevitably lose geometrical structure information during the feature channel generation process, resulting in edge detail mismatches. In this paper, the Motif Cha}nnel Attention Stereo Matching Network (MoCha-Stereo) is designed to address this problem. We provide the Motif Channel Correlation Volume (MCCV) to determine more accurate edge matching costs. MCCV is achieved by projecting motif channels, which capture common geometric structures in feature channels, onto feature maps and cost volumes. In addition, edge variations in %potential feature channels of the reconstruction error map also affect details matching, we propose the Reconstruction Error Motif Penalty (REMP) module to further refine the full-resolution disparity estimation. REMP integrates the frequency information of typical channel features from the reconstruction error. MoCha-Stereo ranks 1st on the KITTI-2015 and KITTI-2012 Reflective leaderboards. Our structure also shows excellent performance in Multi-View Stereo. Code is avaliable at https://github.com/ZYangChen/MoCha-Stereo.
* Accepted to CVPR 2024
HAMIL: Hierarchical Aggregation-Based Multi-Instance Learning for Microscopy Image Classification
Mar 17, 2021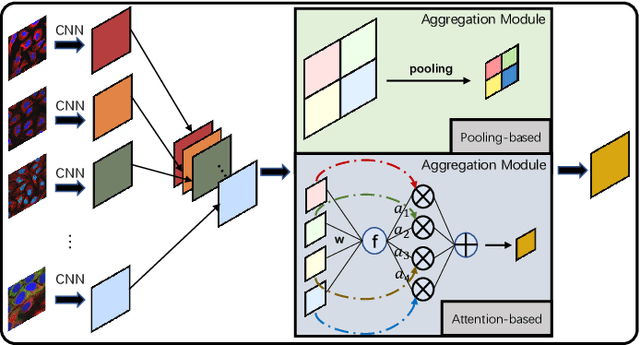
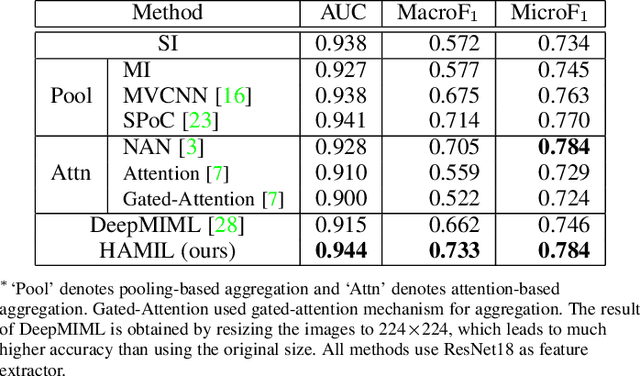
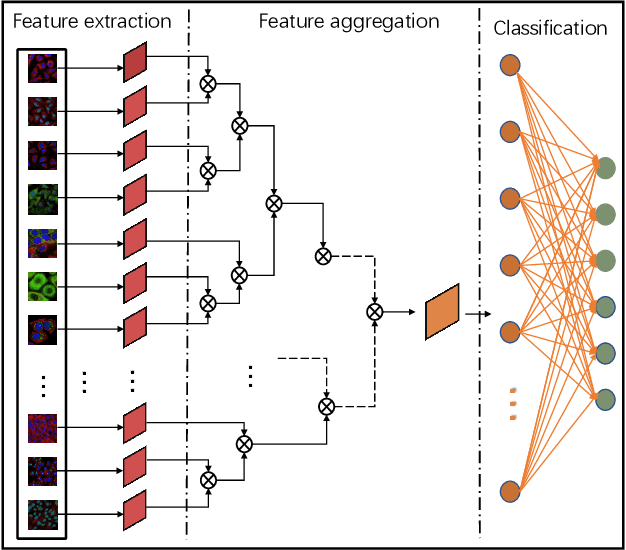
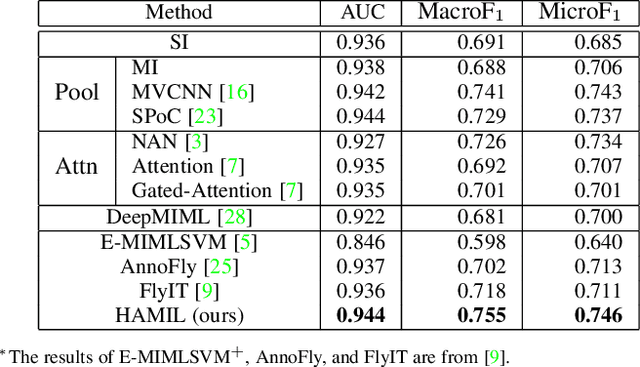
Abstract:Multi-instance learning is common for computer vision tasks, especially in biomedical image processing. Traditional methods for multi-instance learning focus on designing feature aggregation methods and multi-instance classifiers, where the aggregation operation is performed either in feature extraction or learning phase. As deep neural networks (DNNs) achieve great success in image processing via automatic feature learning, certain feature aggregation mechanisms need to be incorporated into common DNN architecture for multi-instance learning. Moreover, flexibility and reliability are crucial considerations to deal with varying quality and number of instances. In this study, we propose a hierarchical aggregation network for multi-instance learning, called HAMIL. The hierarchical aggregation protocol enables feature fusion in a defined order, and the simple convolutional aggregation units lead to an efficient and flexible architecture. We assess the model performance on two microscopy image classification tasks, namely protein subcellular localization using immunofluorescence images and gene annotation using spatial gene expression images. The experimental results show that HAMIL outperforms the state-of-the-art feature aggregation methods and the existing models for addressing these two tasks. The visualization analyses also demonstrate the ability of HAMIL to focus on high-quality instances.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge