Thomas Helmuth
Solving Novel Program Synthesis Problems with Genetic Programming using Parametric Polymorphism
Jun 08, 2023Abstract:Contemporary genetic programming (GP) systems for general program synthesis have been primarily concerned with evolving programs that can manipulate values from a standard set of primitive data types and simple indexed data structures. In contrast, human programmers do not limit themselves to a small finite set of data types and use polymorphism to express an unbounded number of types including nested data structures, product types, and generic functions. Code-building Genetic Programming (CBGP) is a recently introduced method that compiles type-safe programs from linear genomes using stack-based compilation and a formal type system. Although prior work with CBGP has shown initial demonstrations of polymorphism inside evolved programs, we have provided a deeper exploration of these capabilities through the evolution of programs which make use of generic data types such as key-value maps, tuples, and sets, as well as higher order functions and functions with polymorphic type signatures. In our experiments, CBGP is able to solve problems with all of these properties, where every other GP system that we know of has restrictions that make it unable to even consider problems with these properties. This demonstration provides a significant step towards fully aligning the expressiveness of GP to real world programming.
A Static Analysis of Informed Down-Samples
Apr 17, 2023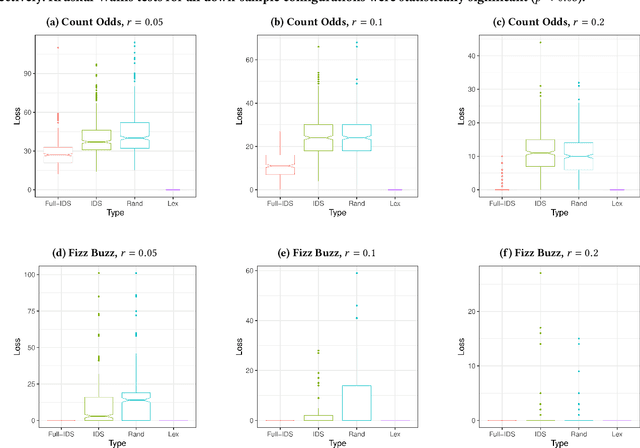
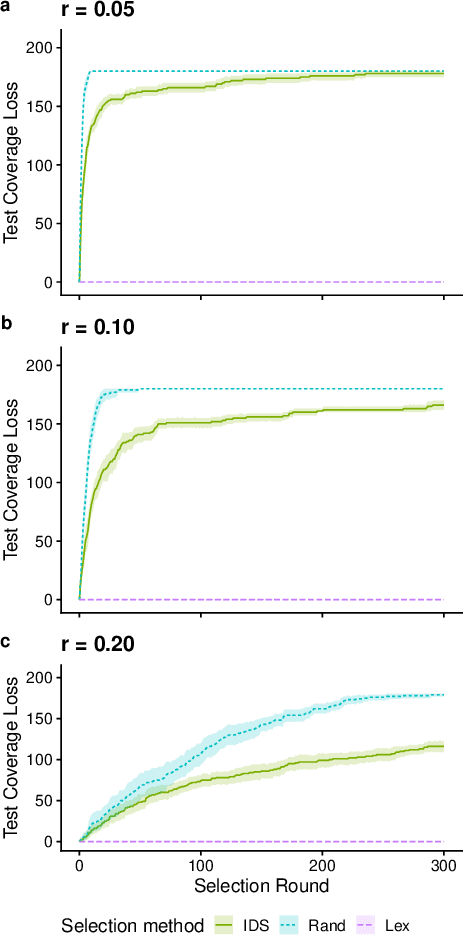
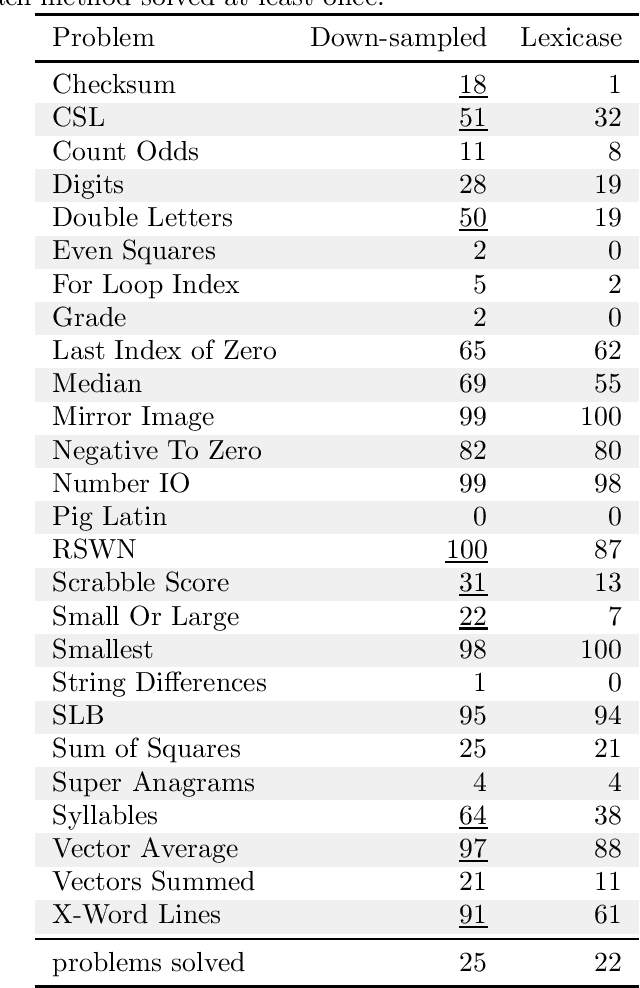
Abstract:We present an analysis of the loss of population-level test coverage induced by different down-sampling strategies when combined with lexicase selection. We study recorded populations from the first generation of genetic programming runs, as well as entirely synthetic populations. Our findings verify the hypothesis that informed down-sampling better maintains population-level test coverage when compared to random down-sampling. Additionally, we show that both forms of down-sampling cause greater test coverage loss than standard lexicase selection with no down-sampling. However, given more information about the population, we found that informed down-sampling can further reduce its test coverage loss. We also recommend wider adoption of the static population analyses we present in this work.
Analyzing the Interaction Between Down-Sampling and Selection
Apr 14, 2023Abstract:Genetic programming systems often use large training sets to evaluate the quality of candidate solutions for selection. However, evaluating populations on large training sets can be computationally expensive. Down-sampling training sets has long been used to decrease the computational cost of evaluation in a wide range of application domains. Indeed, recent studies have shown that both random and informed down-sampling can substantially improve problem-solving success for GP systems that use the lexicase parent selection algorithm. We use the PushGP framework to experimentally test whether these down-sampling techniques can also improve problem-solving success in the context of two other commonly used selection methods, fitness-proportionate and tournament selection, across eight GP problems (four program synthesis and four symbolic regression). We verified that down-sampling can benefit the problem-solving success of both fitness-proportionate and tournament selection. However, the number of problems wherein down-sampling improved problem-solving success varied by selection scheme, suggesting that the impact of down-sampling depends both on the problem and choice of selection scheme. Surprisingly, we found that down-sampling was most consistently beneficial when combined with lexicase selection as compared to tournament and fitness-proportionate selection. Overall, our results suggest that down-sampling should be considered more often when solving test-based GP problems.
Informed Down-Sampled Lexicase Selection: Identifying productive training cases for efficient problem solving
Jan 04, 2023
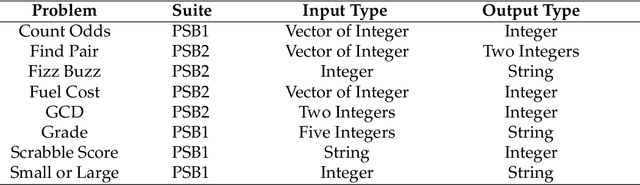


Abstract:Genetic Programming (GP) often uses large training sets and requires all individuals to be evaluated on all training cases during selection. Random down-sampled lexicase selection evaluates individuals on only a random subset of the training cases allowing for more individuals to be explored with the same amount of program executions. However, creating a down-sample randomly might exclude important cases from the current down-sample for a number of generations, while cases that measure the same behavior (synonymous cases) may be overused despite their redundancy. In this work, we introduce Informed Down-Sampled Lexicase Selection. This method leverages population statistics to build down-samples that contain more distinct and therefore informative training cases. Through an empirical investigation across two different GP systems (PushGP and Grammar-Guided GP), we find that informed down-sampling significantly outperforms random down-sampling on a set of contemporary program synthesis benchmark problems. Through an analysis of the created down-samples, we find that important training cases are included in the down-sample consistently across independent evolutionary runs and systems. We hypothesize that this improvement can be attributed to the ability of Informed Down-Sampled Lexicase Selection to maintain more specialist individuals over the course of evolution, while also benefiting from reduced per-evaluation costs.
Lexicase Selection at Scale
Aug 23, 2022
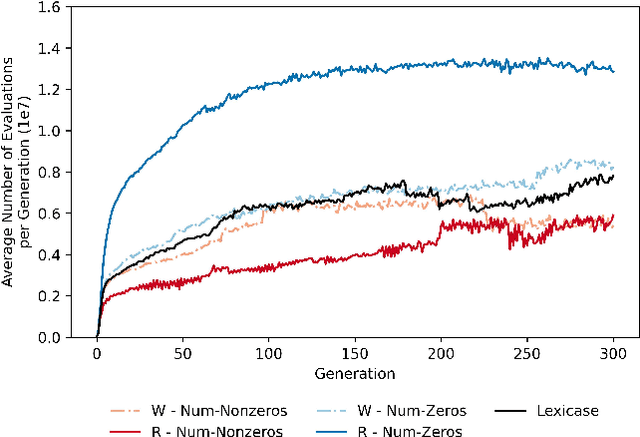

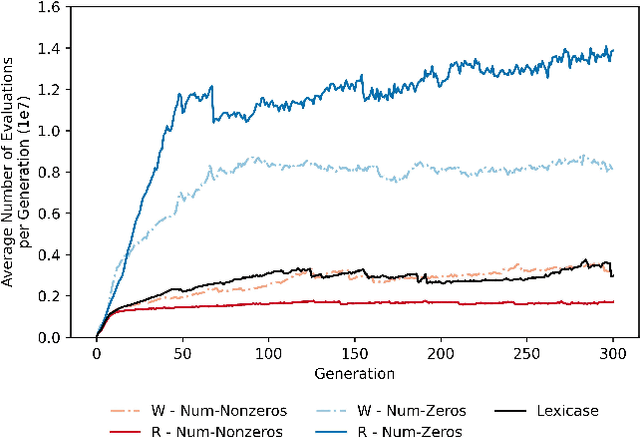
Abstract:Lexicase selection is a semantic-aware parent selection method, which assesses individual test cases in a randomly-shuffled data stream. It has demonstrated success in multiple research areas including genetic programming, genetic algorithms, and more recently symbolic regression and deep learning. One potential drawback of lexicase selection and its variants is that the selection procedure requires evaluating training cases in a single data stream, making it difficult to handle tasks where the evaluation is computationally heavy or the dataset is large-scale, e.g., deep learning. In this work, we investigate how the weighted shuffle methods can be employed to improve the efficiency of lexicase selection. We propose a novel method, fast lexicase selection, which incorporates lexicase selection and weighted shuffle with partial evaluation. Experiments on both classic genetic programming and deep learning tasks indicate that the proposed method can significantly reduce the number of evaluation steps needed for lexicase selection to select an individual, improving its efficiency while maintaining the performance.
Functional Code Building Genetic Programming
Jun 09, 2022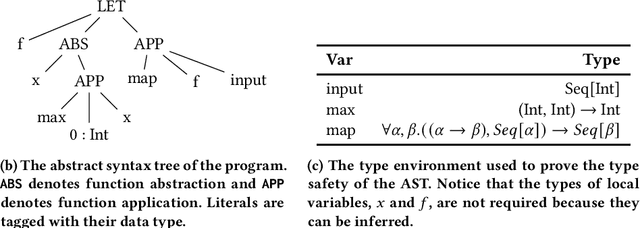
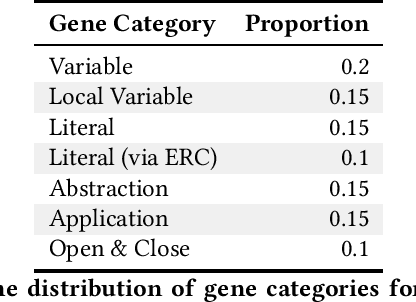
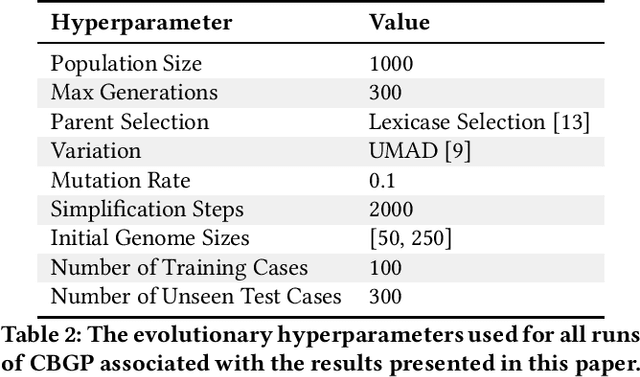

Abstract:General program synthesis has become an important application area for genetic programming (GP), and for artificial intelligence more generally. Code Building Genetic Programming (CBGP) is a recently introduced GP method for general program synthesis that leverages reflection and first class specifications to support the evolution of programs that may use arbitrary data types, polymorphism, and functions drawn from existing codebases. However, neither a formal description nor a thorough benchmarking of CBGP have yet been reported. In this work, we formalize the method of CBGP using algorithms from type theory. Specially, we show that a functional programming language and a Hindley-Milner type system can be used to evolve type-safe programs using the process abstractly described in the original CBGP paper. Furthermore, we perform a comprehensive analysis of the search performance of this functional variant of CBGP compared to other contemporary GP program synthesis methods.
The Environmental Discontinuity Hypothesis for Down-Sampled Lexicase Selection
May 31, 2022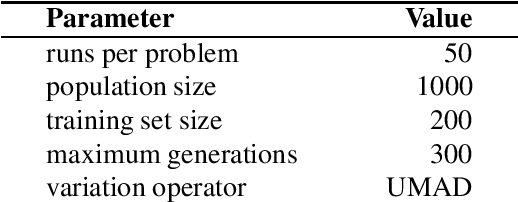
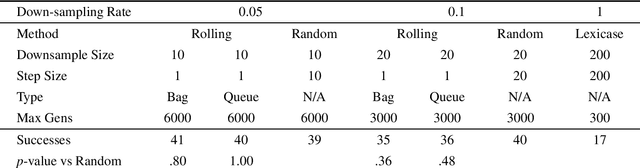
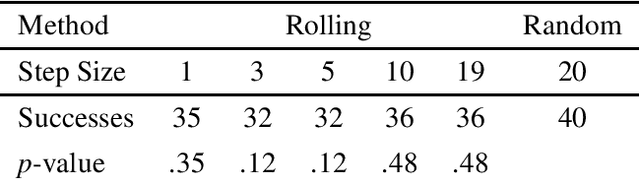
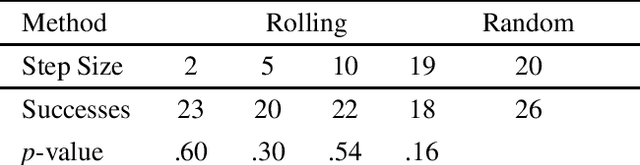
Abstract:Down-sampling training data has long been shown to improve the generalization performance of a wide range of machine learning systems. Recently, down-sampling has proved effective in genetic programming (GP) runs that utilize the lexicase parent selection technique. Although this down-sampling procedure has been shown to significantly improve performance across a variety of problems, it does not seem to do so due to encouraging adaptability through environmental change. We hypothesize that the random sampling that is performed every generation causes discontinuities that result in the population being unable to adapt to the shifting environment. We investigate modifications to down-sampled lexicase selection in hopes of promoting incremental environmental change to scaffold evolution by reducing the amount of jarring discontinuities between the environments of successive generations. In our empirical studies, we find that forcing incremental environmental change is not significantly better for evolving solutions to program synthesis problems than simple random down-sampling. In response to this, we attempt to exacerbate the hypothesized prevalence of discontinuities by using only disjoint down-samples to see if it hinders performance. We find that this also does not significantly differ from the performance of regular random down-sampling. These negative results raise new questions about the ways in which the composition of sub-samples, which may include synonymous cases, may be expected to influence the performance of machine learning systems that use down-sampling.
Population Diversity Leads to Short Running Times of Lexicase Selection
Apr 13, 2022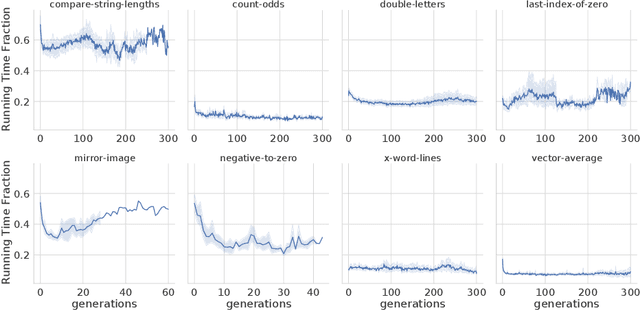
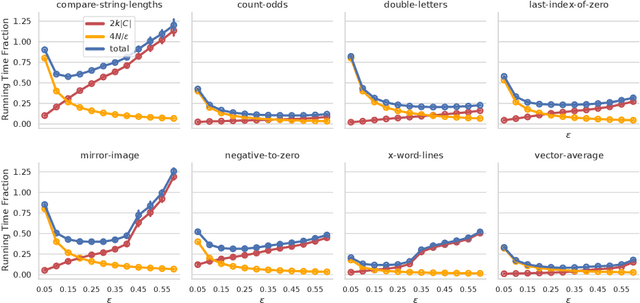
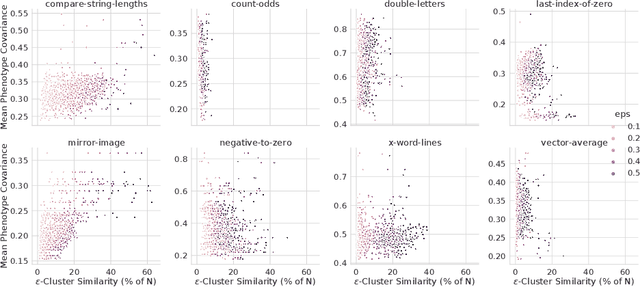
Abstract:In this paper we investigate why the running time of lexicase parent selection is empirically much lower than its worst-case bound of O(N*C). We define a measure of population diversity and prove that high diversity leads to low running times O(N + C) of lexicase selection. We then show empirically that genetic programming populations evolved under lexicase selection are diverse for several program synthesis problems, and explore the resulting differences in running time bounds.
PSB2: The Second Program Synthesis Benchmark Suite
Jun 10, 2021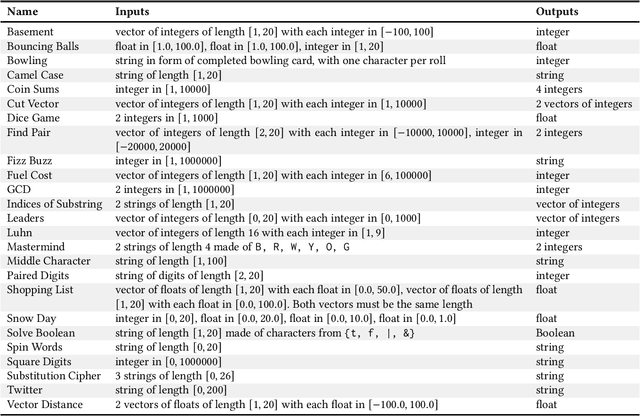
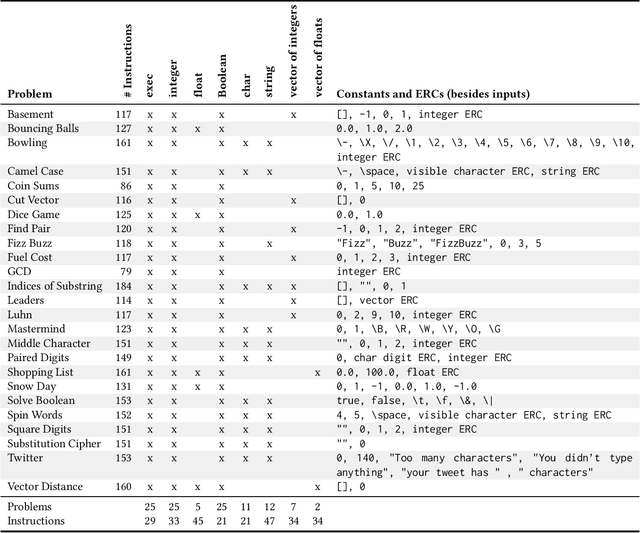
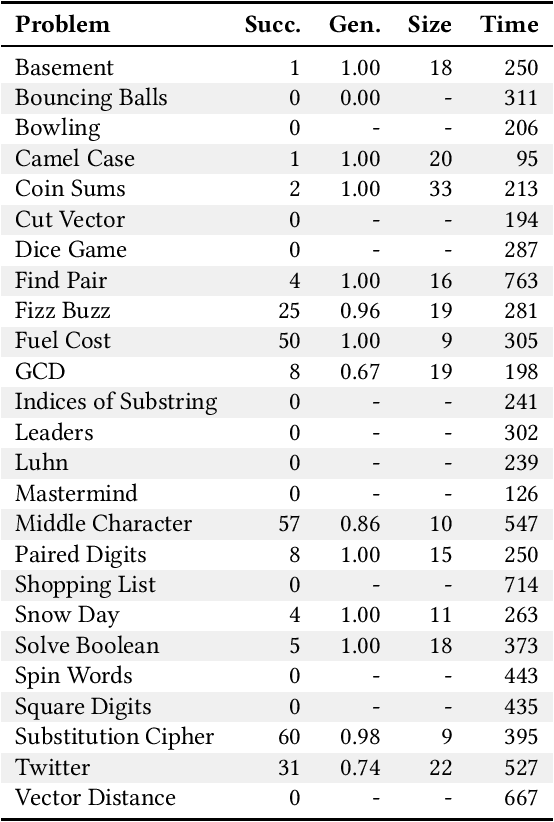
Abstract:For the past six years, researchers in genetic programming and other program synthesis disciplines have used the General Program Synthesis Benchmark Suite to benchmark many aspects of automatic program synthesis systems. These problems have been used to make notable progress toward the goal of general program synthesis: automatically creating the types of software that human programmers code. Many of the systems that have attempted the problems in the original benchmark suite have used it to demonstrate performance improvements granted through new techniques. Over time, the suite has gradually become outdated, hindering the accurate measurement of further improvements. The field needs a new set of more difficult benchmark problems to move beyond what was previously possible. In this paper, we describe the 25 new general program synthesis benchmark problems that make up PSB2, a new benchmark suite. These problems are curated from a variety of sources, including programming katas and college courses. We selected these problems to be more difficult than those in the original suite, and give results using PushGP showing this increase in difficulty. These new problems give plenty of room for improvement, pointing the way for the next six or more years of general program synthesis research.
Problem-solving benefits of down-sampled lexicase selection
Jun 10, 2021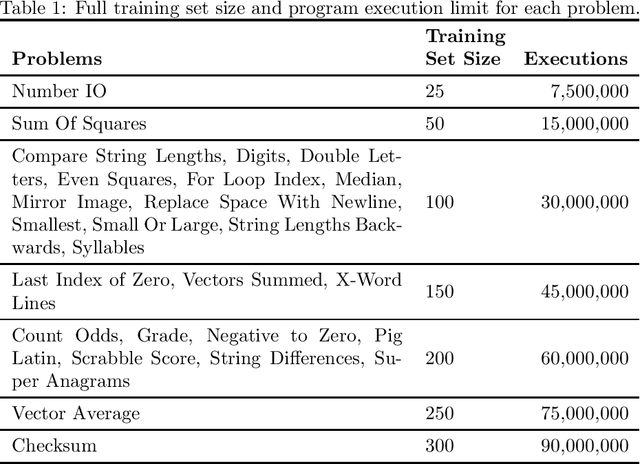

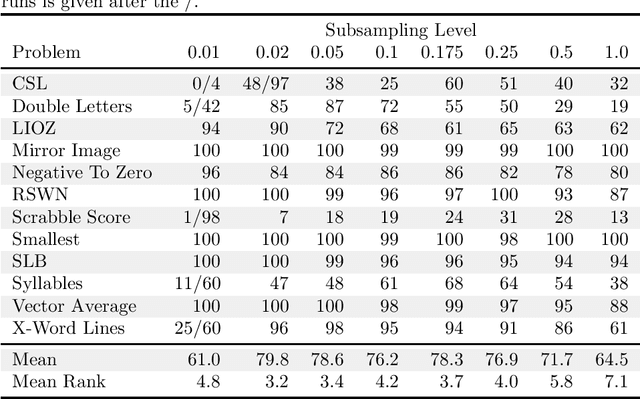
Abstract:In genetic programming, an evolutionary method for producing computer programs that solve specified computational problems, parent selection is ordinarily based on aggregate measures of performance across an entire training set. Lexicase selection, by contrast, selects on the basis of performance on random sequences of training cases; this has been shown to enhance problem-solving power in many circumstances. Lexicase selection can also be seen as better reflecting biological evolution, by modeling sequences of challenges that organisms face over their lifetimes. Recent work has demonstrated that the advantages of lexicase selection can be amplified by down-sampling, meaning that only a random subsample of the training cases is used each generation. This can be seen as modeling the fact that individual organisms encounter only subsets of the possible environments, and that environments change over time. Here we provide the most extensive benchmarking of down-sampled lexicase selection to date, showing that its benefits hold up to increased scrutiny. The reasons that down-sampling helps, however, are not yet fully understood. Hypotheses include that down-sampling allows for more generations to be processed with the same budget of program evaluations; that the variation of training data across generations acts as a changing environment, encouraging adaptation; or that it reduces overfitting, leading to more general solutions. We systematically evaluate these hypotheses, finding evidence against all three, and instead draw the conclusion that down-sampled lexicase selection's main benefit stems from the fact that it allows the evolutionary process to examine more individuals within the same computational budget, even though each individual is examined less completely.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge