Charles Ofria
Michigan State University
ECLIPSE: An Evolutionary Computation Library for Instrumentation Prototyping in Scientific Engineering
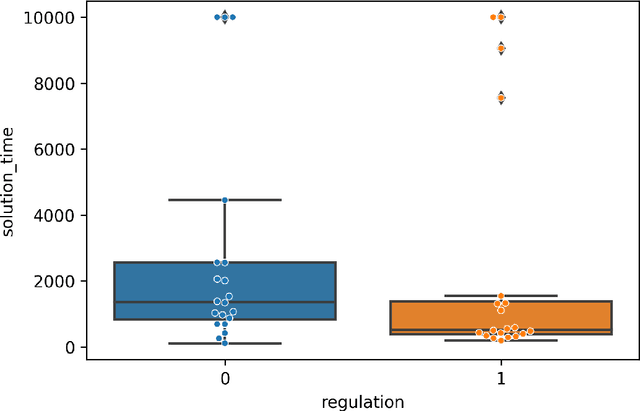
Jan 08, 2026Abstract:Designing scientific instrumentation often requires exploring large, highly constrained design spaces using computationally expensive physics simulations. These simulators pose substantial challenges for integrating evolutionary computation (EC) into scientific design workflows. Evolutionary computation typically requires numerous design evaluations, making the integration of slow, low-throughput simulators particularly challenging, as they are optimized for accuracy and ease of use rather than throughput. We present ECLIPSE, an evolutionary computation framework built to interface directly with complex, domain-specific simulation tools while supporting flexible geometric and parametric representations of scientific hardware. ECLIPSE provides a modular architecture consisting of (1) Individuals, which encode hardware designs using domain-aware, physically constrained representations; (2) Evaluators, which prepare simulation inputs, invoke external simulators, and translate the simulator's outputs into fitness measures; and (3) Evolvers, which implement EC algorithms suitable for high-cost, limited-throughput environments. We demonstrate the utility of ECLIPSE across several active space-science applications, including evolved 3D antennas and spacecraft geometries optimized for drag reduction in very low Earth orbit. We further discuss the practical challenges encountered when coupling EC with scientific simulation workflows, including interoperability constraints, parallelization limits, and extreme evaluation costs, and outline ongoing efforts to combat these challenges. ECLIPSE enables interdisciplinary teams of physicists, engineers, and EC researchers to collaboratively explore unconventional designs for scientific hardware while leveraging existing domain-specific simulation software.
Case Study of Novelty, Complexity, and Adaptation in a Multicellular System
May 12, 2024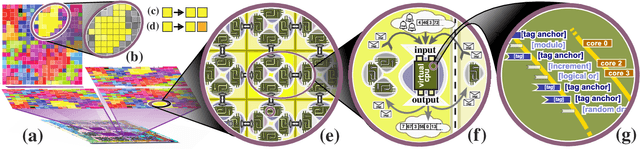
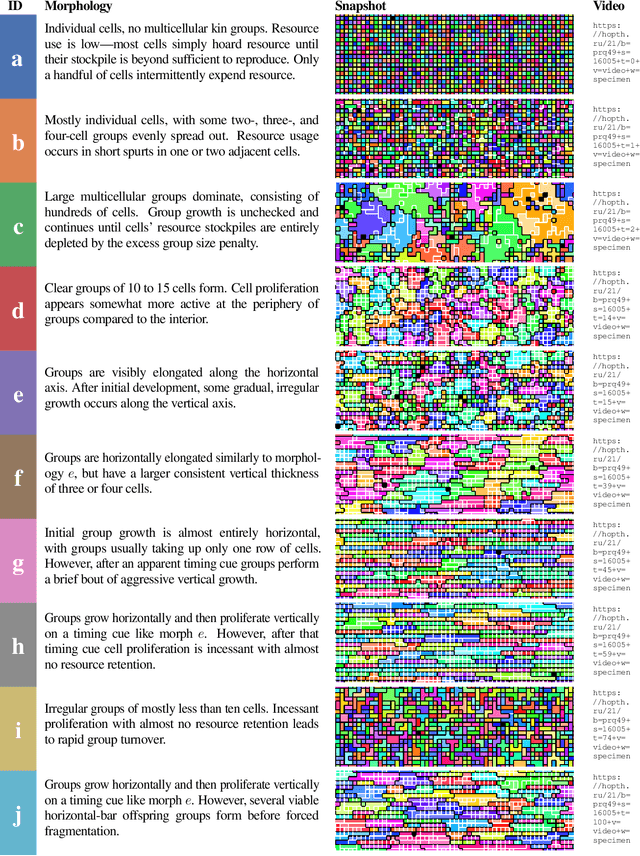
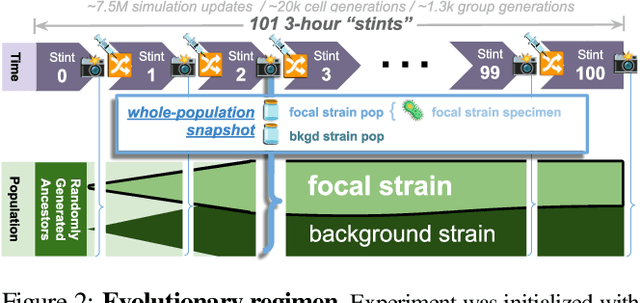
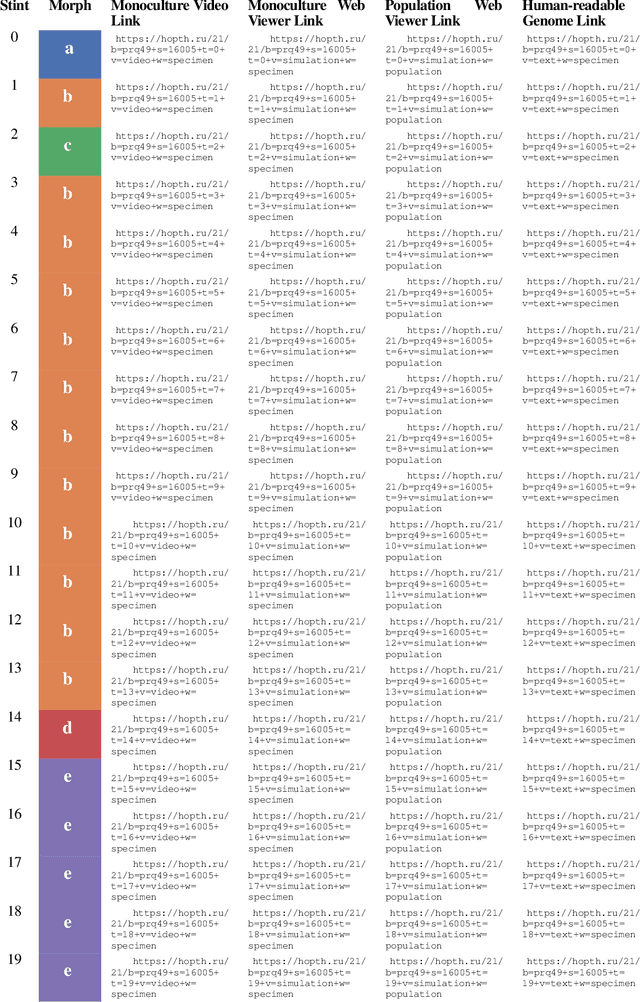
Abstract:Continuing generation of novelty, complexity, and adaptation are well-established as core aspects of open-ended evolution. However, it has yet to be firmly established to what extent these phenomena are coupled and by what means they interact. In this work, we track the co-evolution of novelty, complexity, and adaptation in a case study from the DISHTINY simulation system, which is designed to study the evolution of digital multicellularity. In this case study, we describe ten qualitatively distinct multicellular morphologies, several of which exhibit asymmetrical growth and distinct life stages. We contextualize the evolutionary history of these morphologies with measurements of complexity and adaptation. Our case study suggests a loose -- sometimes divergent -- relationship can exist among novelty, complexity, and adaptation.
Informed Down-Sampled Lexicase Selection: Identifying productive training cases for efficient problem solving
Jan 04, 2023


Abstract:Genetic Programming (GP) often uses large training sets and requires all individuals to be evaluated on all training cases during selection. Random down-sampled lexicase selection evaluates individuals on only a random subset of the training cases allowing for more individuals to be explored with the same amount of program executions. However, creating a down-sample randomly might exclude important cases from the current down-sample for a number of generations, while cases that measure the same behavior (synonymous cases) may be overused despite their redundancy. In this work, we introduce Informed Down-Sampled Lexicase Selection. This method leverages population statistics to build down-samples that contain more distinct and therefore informative training cases. Through an empirical investigation across two different GP systems (PushGP and Grammar-Guided GP), we find that informed down-sampling significantly outperforms random down-sampling on a set of contemporary program synthesis benchmark problems. Through an analysis of the created down-samples, we find that important training cases are included in the down-sample consistently across independent evolutionary runs and systems. We hypothesize that this improvement can be attributed to the ability of Informed Down-Sampled Lexicase Selection to maintain more specialist individuals over the course of evolution, while also benefiting from reduced per-evaluation costs.
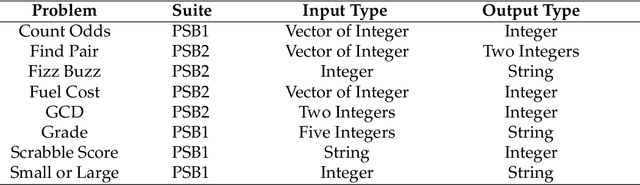
A suite of diagnostic metrics for characterizing selection schemes
Apr 29, 2022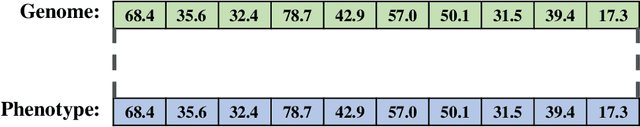
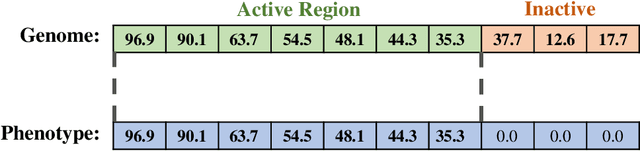
Abstract:Evolutionary algorithms are effective general-purpose techniques for solving optimization problems. Understanding how each component of an evolutionary algorithm influences its problem-solving success improves our ability to target particular problem domains. Our work focuses on evaluating selection schemes, which choose individuals to contribute genetic material to the next generation. We introduce four diagnostic search spaces for testing the strengths and weaknesses of selection schemes: the exploitation rate diagnostic, ordered exploitation rate diagnostic, contradictory objectives diagnostic, and the multi-path exploration diagnostic. Each diagnostic is handcrafted to isolate and measure the relative exploitation and exploration characteristics of selection schemes. In this study, we use our diagnostics to evaluate six population selection methods: truncation selection, tournament selection, fitness sharing, lexicase selection, nondominated sorting, and novelty search. Expectedly, tournament and truncation selection excelled in gradient exploitation but poorly explored search spaces, and novelty search excelled at exploration but failed to exploit fitness gradients. Fitness sharing performed poorly across all diagnostics, suggesting poor overall exploitation and exploration abilities. Nondominated sorting was best for maintaining populations comprised of individuals with different trade-offs of multiple objectives, but struggled to effectively exploit fitness gradients. Lexicase selection balanced search space exploration with exploitation, generally performing well across diagnostics. Our work demonstrates the value of diagnostic search spaces for building a deeper understanding of selection schemes, which can then be used to improve or develop new selection methods.
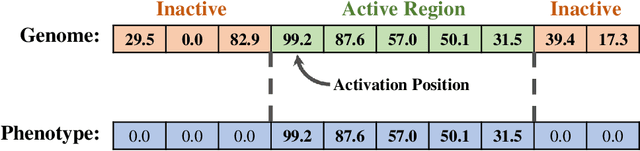
Matchmaker, Matchmaker, Make Me a Match: Geometric, Variational, and Evolutionary Implications of Criteria for Tag Affinity
Aug 10, 2021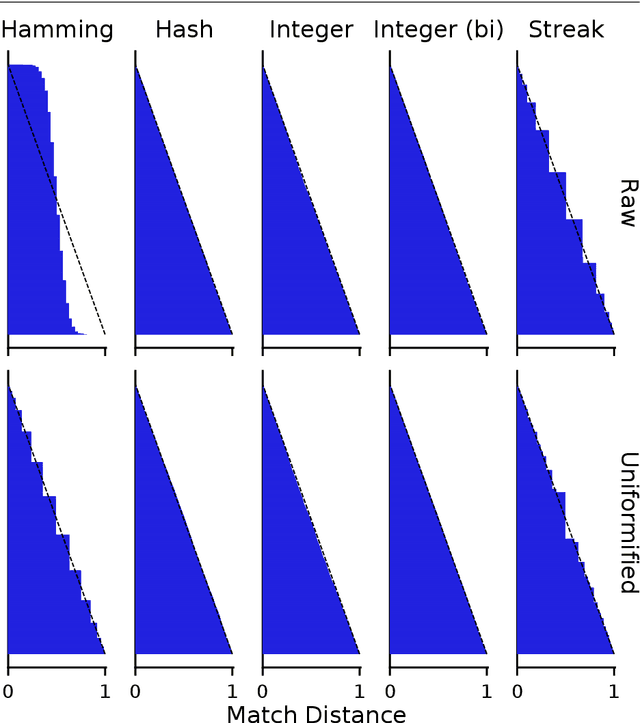

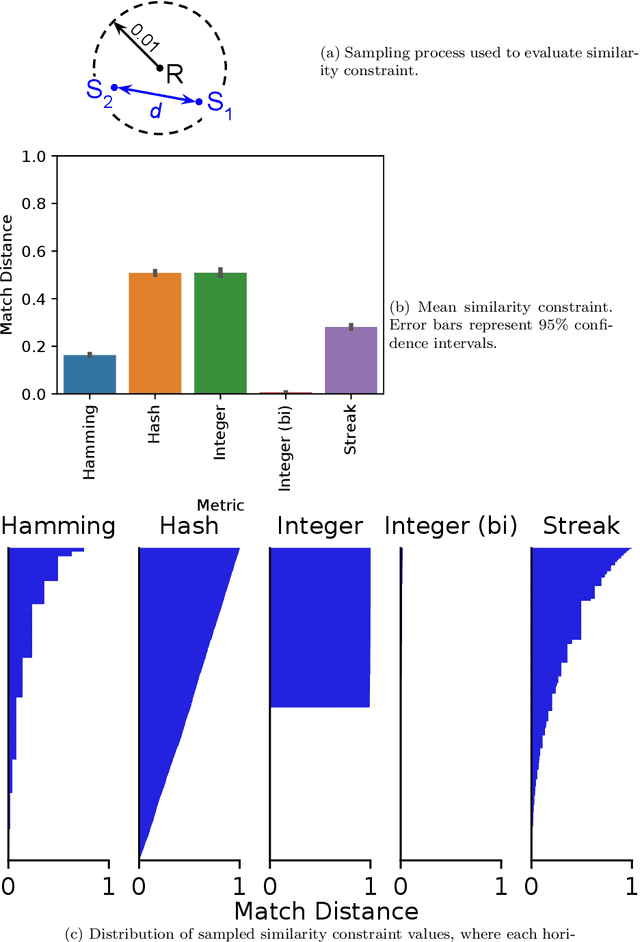
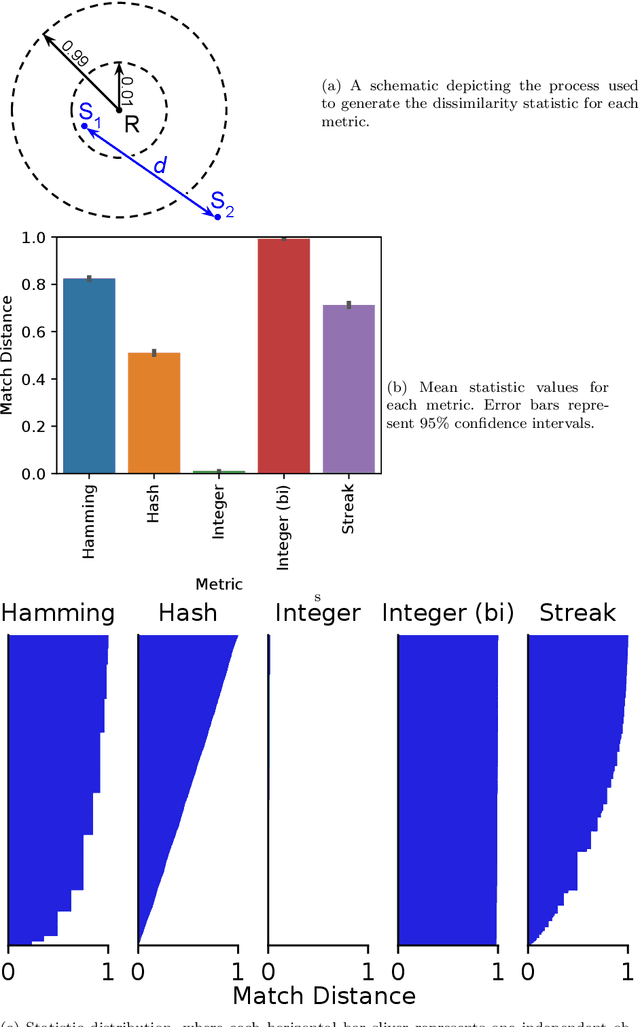
Abstract:Genetic programming and artificial life systems commonly employ tag-matching schemes to determine interactions between model components. However, the implications of criteria used to determine affinity between tags with respect to constraints on emergent connectivity, canalization of changes to connectivity under mutation, and evolutionary dynamics have not been considered. We highlight differences between tag-matching criteria with respect to geometric constraint and variation generated under mutation. We find that tag-matching criteria can influence the rate of adaptive evolution and the quality of evolved solutions. Better understanding of the geometric, variational, and evolutionary properties of tag-matching criteria will facilitate more effective incorporation of tag matching into genetic programming and artificial life systems. By showing that tag-matching criteria influence connectivity patterns and evolutionary dynamics, our findings also raise fundamental questions about the properties of tag-matching systems in nature.
SignalGP-Lite: Event Driven Genetic Programming Library for Large-Scale Artificial Life Applications
Aug 01, 2021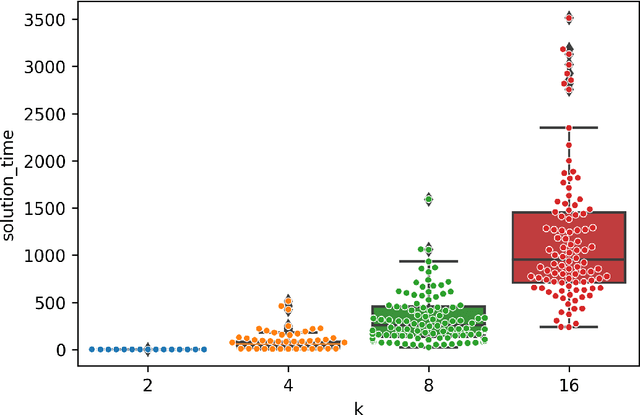

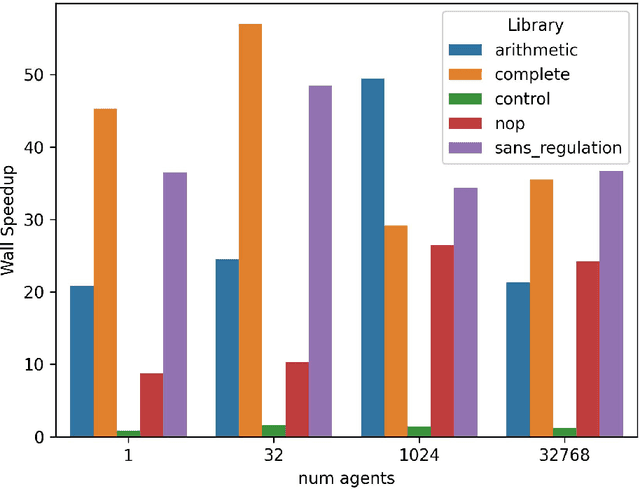
Abstract:Event-driven genetic programming representations have been shown to outperform traditional imperative representations on interaction-intensive problems. The event-driven approach organizes genome content into modules that are triggered in response to environmental signals, simplifying simulation design and implementation. Existing work developing event-driven genetic programming methodology has largely used the SignalGP library, which caters to traditional program synthesis applications. The SignalGP-Lite library enables larger-scale artificial life experiments with streamlined agents by reducing control flow overhead and trading run-time flexibility for better performance due to compile-time configuration. Here, we report benchmarking experiments that show an 8x to 30x speedup. We also report solution quality equivalent to SignalGP on two benchmark problems originally developed to test the ability of evolved programs to respond to a large number of signals and to modulate signal response based on context.
An Exploration of Exploration: Measuring the ability of lexicase selection to find obscure pathways to optimality
Jul 26, 2021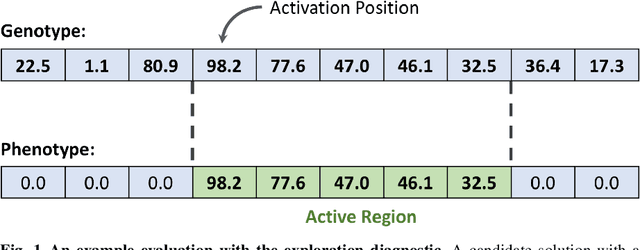
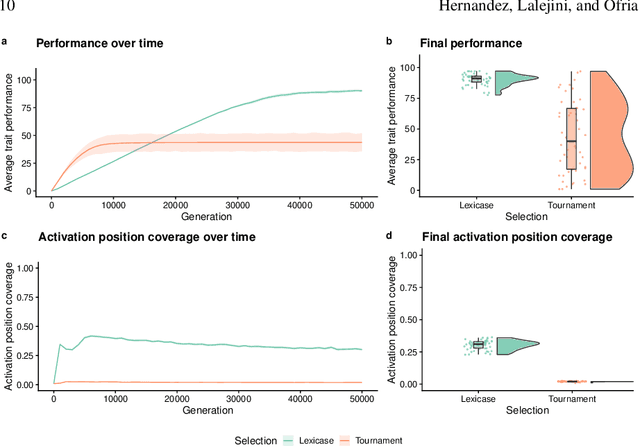
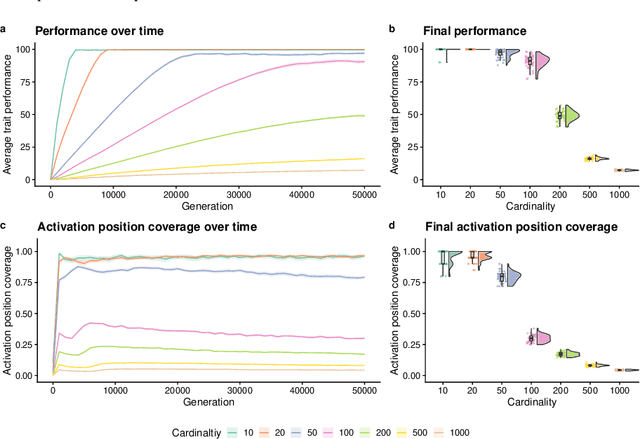
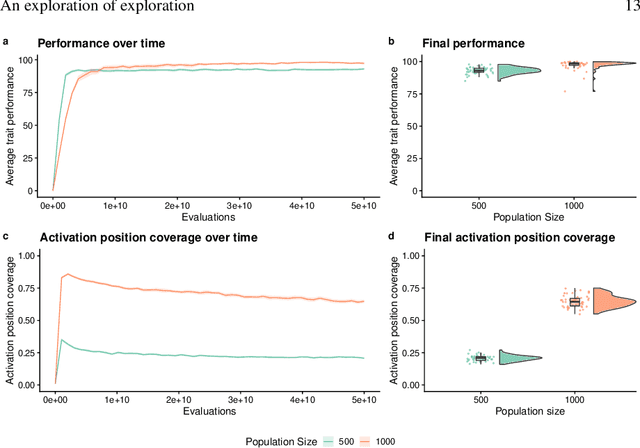
Abstract:Parent selection algorithms (selection schemes) steer populations through a problem's search space, often trading off between exploitation and exploration. Understanding how selection schemes affect exploitation and exploration within a search space is crucial to tackling increasingly challenging problems. Here, we introduce an "exploration diagnostic" that diagnoses a selection scheme's capacity for search space exploration. We use our exploration diagnostic to investigate the exploratory capacity of lexicase selection and several of its variants: epsilon lexicase, down-sampled lexicase, cohort lexicase, and novelty-lexicase. We verify that lexicase selection out-explores tournament selection, and we show that lexicase selection's exploratory capacity can be sensitive to the ratio between population size and the number of test cases used for evaluating candidate solutions. Additionally, we find that relaxing lexicase's elitism with epsilon lexicase can further improve exploration. Both down-sampling and cohort lexicase -- two techniques for applying random subsampling to test cases -- degrade lexicase's exploratory capacity; however, we find that cohort partitioning better preserves lexicase's exploratory capacity than down-sampling. Finally, we find evidence that novelty-lexicase's addition of novelty test cases can degrade lexicase's capacity for exploration. Overall, our findings provide hypotheses for further exploration and actionable insights and recommendations for using lexicase selection. Additionally, this work demonstrates the value of selection scheme diagnostics as a complement to more conventional benchmarking approaches to selection scheme analysis.
Exploring Evolved Multicellular Life Histories in a Open-Ended Digital Evolution System
Apr 20, 2021

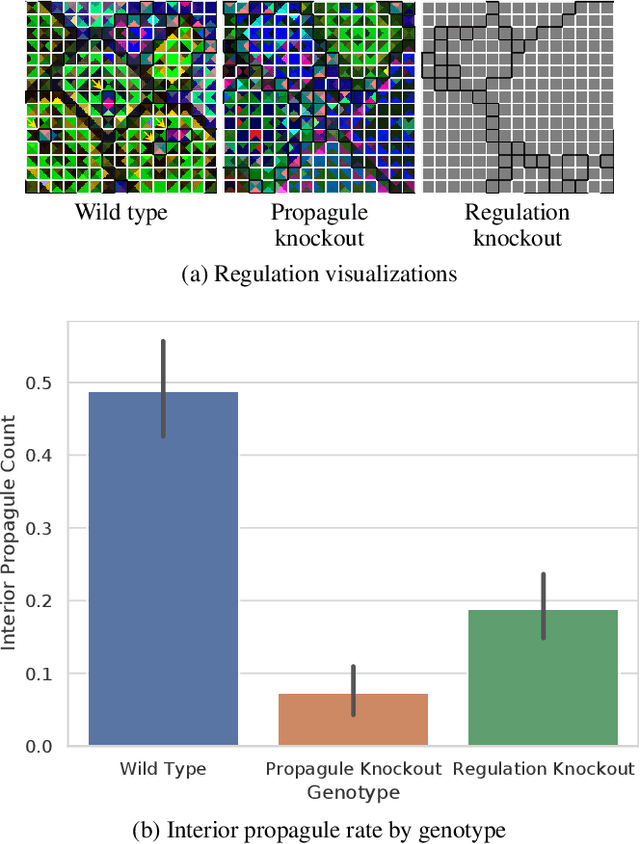
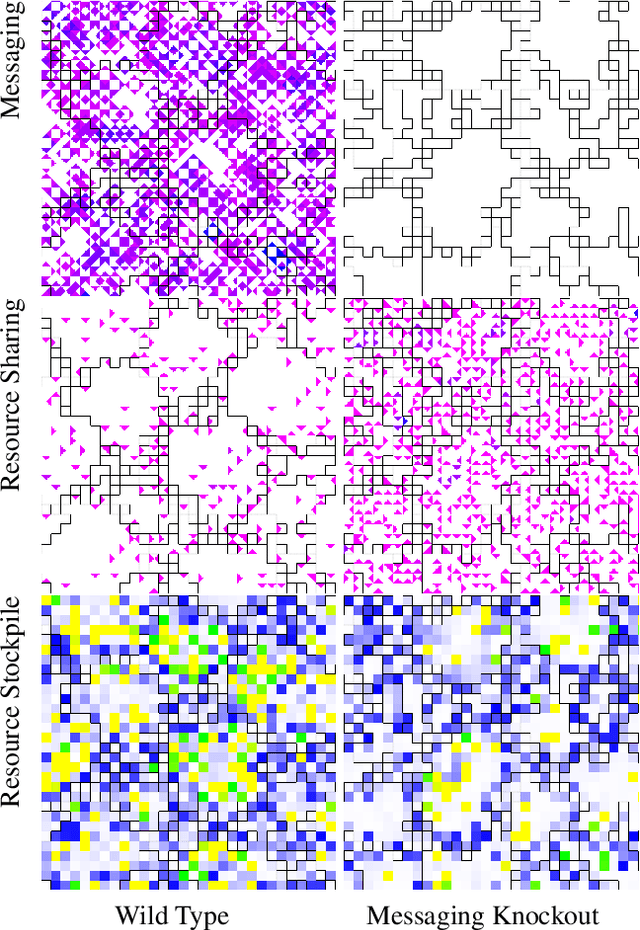
Abstract:Evolutionary transitions occur when previously-independent replicating entities unite to form more complex individuals. Such transitions have profoundly shaped natural evolutionary history and occur in two forms: fraternal transitions involve lower-level entities that are kin (e.g., transitions to multicellularity or to eusocial colonies), while egalitarian transitions involve unrelated individuals (e.g., the origins of mitochondria). The necessary conditions and evolutionary mechanisms for these transitions to arise continue to be fruitful targets of scientific interest. Here, we examine a range of fraternal transitions in populations of open-ended self-replicating computer programs. These digital cells were allowed to form and replicate kin groups by selectively adjoining or expelling daughter cells. The capability to recognize kin-group membership enabled preferential communication and cooperation between cells. We repeatedly observed group-level traits that are characteristic of a fraternal transition. These included reproductive division of labor, resource sharing within kin groups, resource investment in offspring groups, asymmetrical behaviors mediated by messaging, morphological patterning, and adaptive apoptosis. We report eight case studies from replicates where transitions occurred and explore the diverse range of adaptive evolved multicellular strategies.
Tag-based Genetic Regulation for Genetic Programming
Dec 23, 2020
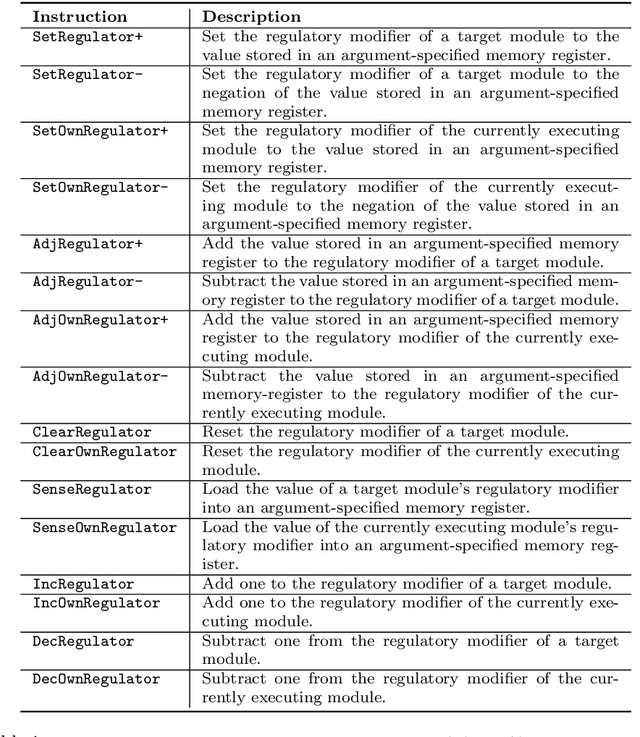
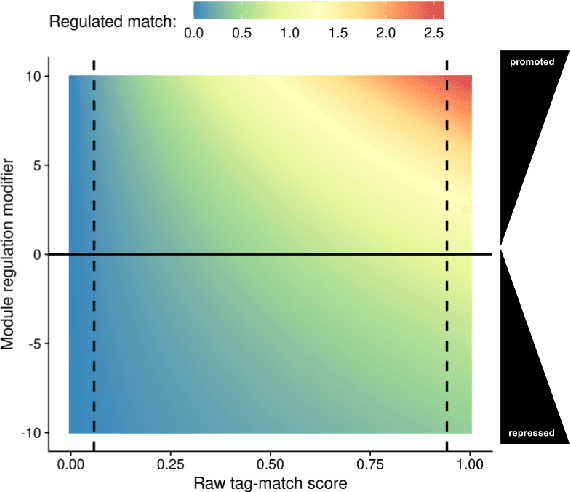

Abstract:We introduce and experimentally demonstrate tag-based genetic regulation, a new genetic programming (GP) technique that allows evolving programs to dynamically adjust which code modules to express. Tags are evolvable labels that provide a flexible mechanism for referring to code modules. Tag-based genetic regulation extends existing tag-based naming schemes to allow programs to "promote" and "repress" code modules. This extension allows evolution to structure a program as a gene regulatory network where program modules are regulated based on instruction executions. We demonstrate the functionality of tag-based regulation on a range of program synthesis problems. We find that tag-based regulation improves problem-solving performance on context-dependent problems; that is, problems where programs must adjust how they respond to current inputs based on prior inputs (i.e., current context). We also observe that our implementation of tag-based genetic regulation can impede adaptive evolution when expected outputs are not context-dependent (i.e., the correct response to a particular input remains static over time). Tag-based genetic regulation broadens our repertoire of techniques for evolving more dynamic genetic programs and can easily be incorporated into existing tag-enabled GP systems.
The Surprising Creativity of Digital Evolution: A Collection of Anecdotes from the Evolutionary Computation and Artificial Life Research Communities
Aug 14, 2018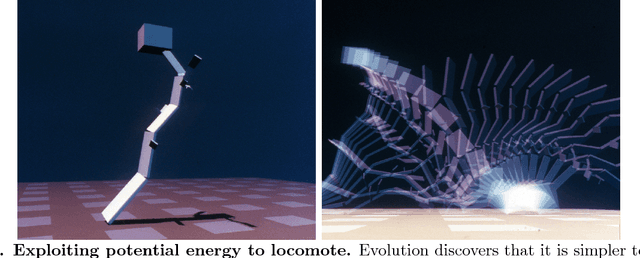
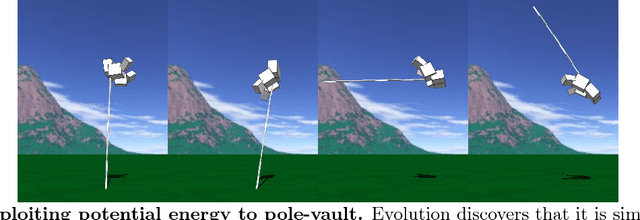
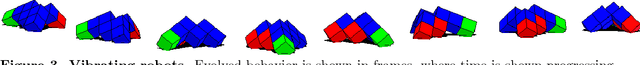

Abstract:Biological evolution provides a creative fount of complex and subtle adaptations, often surprising the scientists who discover them. However, because evolution is an algorithmic process that transcends the substrate in which it occurs, evolution's creativity is not limited to nature. Indeed, many researchers in the field of digital evolution have observed their evolving algorithms and organisms subverting their intentions, exposing unrecognized bugs in their code, producing unexpected adaptations, or exhibiting outcomes uncannily convergent with ones in nature. Such stories routinely reveal creativity by evolution in these digital worlds, but they rarely fit into the standard scientific narrative. Instead they are often treated as mere obstacles to be overcome, rather than results that warrant study in their own right. The stories themselves are traded among researchers through oral tradition, but that mode of information transmission is inefficient and prone to error and outright loss. Moreover, the fact that these stories tend to be shared only among practitioners means that many natural scientists do not realize how interesting and lifelike digital organisms are and how natural their evolution can be. To our knowledge, no collection of such anecdotes has been published before. This paper is the crowd-sourced product of researchers in the fields of artificial life and evolutionary computation who have provided first-hand accounts of such cases. It thus serves as a written, fact-checked collection of scientifically important and even entertaining stories. In doing so we also present here substantial evidence that the existence and importance of evolutionary surprises extends beyond the natural world, and may indeed be a universal property of all complex evolving systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge