Jose Guadalupe Hernandez
GP and LLMs for Program Synthesis: No Clear Winners
Aug 05, 2025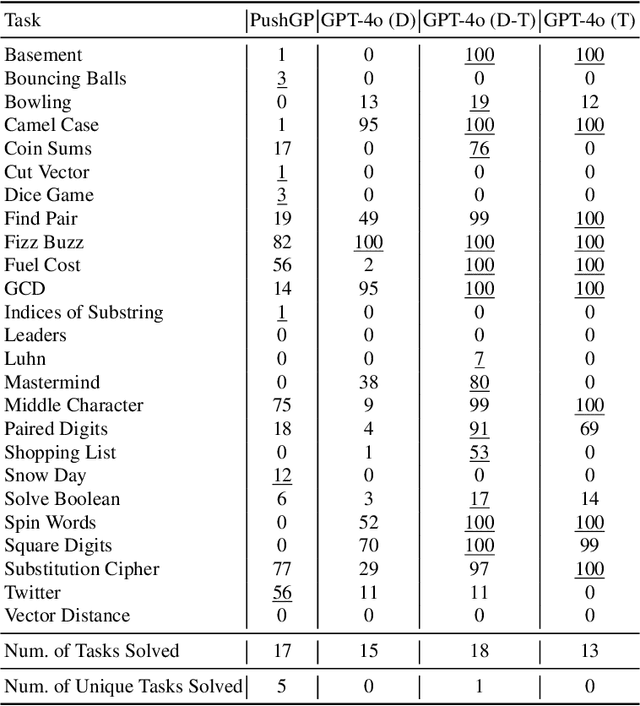
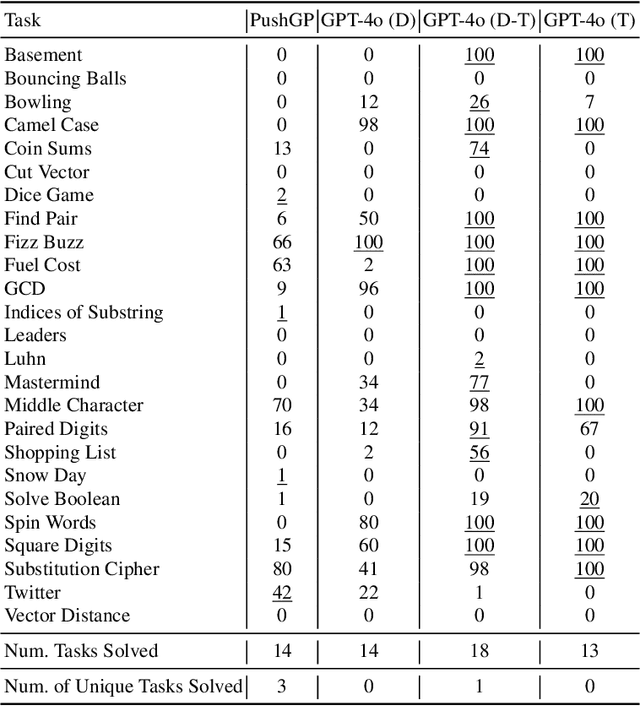
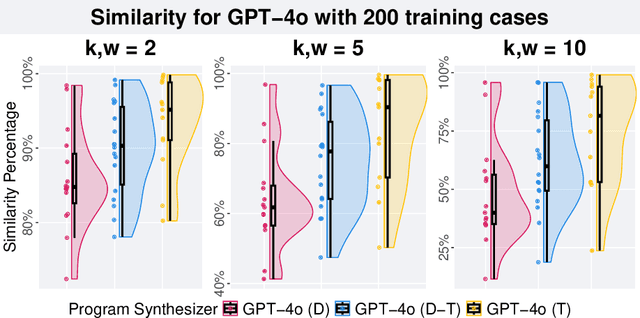
Abstract:Genetic programming (GP) and large language models (LLMs) differ in how program specifications are provided: GP uses input-output examples, and LLMs use text descriptions. In this work, we compared the ability of PushGP and GPT-4o to synthesize computer programs for tasks from the PSB2 benchmark suite. We used three prompt variants with GPT-4o: input-output examples (data-only), textual description of the task (text-only), and a combination of both textual descriptions and input-output examples (data-text). Additionally, we varied the number of input-output examples available for building programs. For each synthesizer and task combination, we compared success rates across all program synthesizers, as well as the similarity between successful GPT-4o synthesized programs. We found that the combination of PushGP and GPT-4o with data-text prompting led to the greatest number of tasks solved (23 of the 25 tasks), even though several tasks were solved exclusively by only one of the two synthesizers. We also observed that PushGP and GPT-4o with data-only prompting solved fewer tasks with the decrease in the training set size, while the remaining synthesizers saw no decrease. We also detected significant differences in similarity between the successful programs synthesized for GPT-4o with text-only and data-only prompting. With there being no dominant program synthesizer, this work highlights the importance of different optimization techniques used by PushGP and LLMs to synthesize programs.
StarBASE-GP: Biologically-Guided Automated Machine Learning for Genotype-to-Phenotype Association Analysis
May 28, 2025Abstract:We present the Star-Based Automated Single-locus and Epistasis analysis tool - Genetic Programming (StarBASE-GP), an automated framework for discovering meaningful genetic variants associated with phenotypic variation in large-scale genomic datasets. StarBASE-GP uses a genetic programming-based multi-objective optimization strategy to evolve machine learning pipelines that simultaneously maximize explanatory power (r2) and minimize pipeline complexity. Biological domain knowledge is integrated at multiple stages, including the use of nine inheritance encoding strategies to model deviations from additivity, a custom linkage disequilibrium pruning node that minimizes redundancy among features, and a dynamic variant recommendation system that prioritizes informative candidates for pipeline inclusion. We evaluate StarBASE-GP on a cohort of Rattus norvegicus (brown rat) to identify variants associated with body mass index, benchmarking its performance against a random baseline and a biologically naive version of the tool. StarBASE-GP consistently evolves Pareto fronts with superior performance, yielding higher accuracy in identifying both ground truth and novel quantitative trait loci, highlighting relevant targets for future validation. By incorporating evolutionary search and relevant biological theory into a flexible automated machine learning framework, StarBASE-GP demonstrates robust potential for advancing variant discovery in complex traits.
Lexicase Selection Parameter Analysis: Varying Population Size and Test Case Redundancy with Diagnostic Metrics
Jul 21, 2024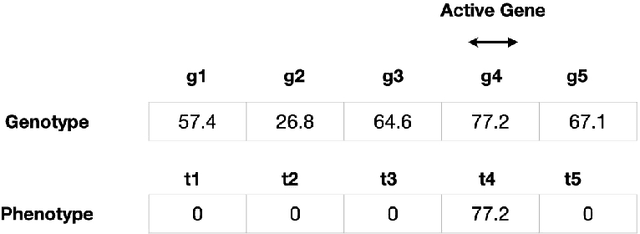
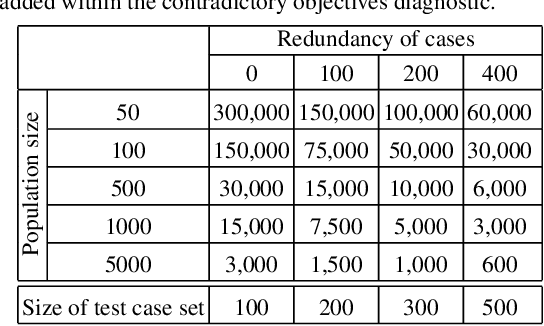
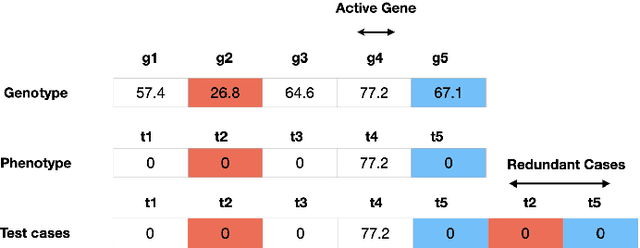
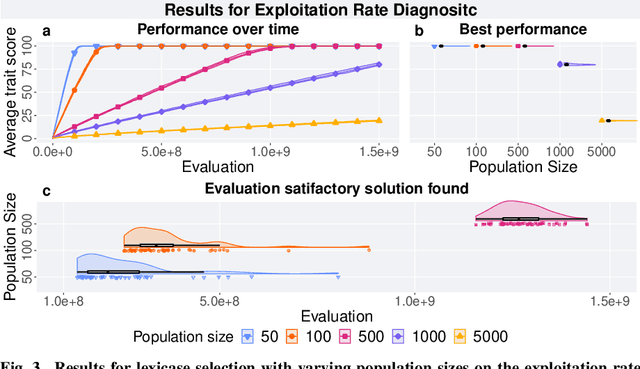
Abstract:Lexicase selection is a successful parent selection method in genetic programming that has outperformed other methods across multiple benchmark suites. Unlike other selection methods that require explicit parameters to function, such as tournament size in tournament selection, lexicase selection does not. However, if evolutionary parameters like population size and number of generations affect the effectiveness of a selection method, then lexicase's performance may also be impacted by these `hidden' parameters. Here, we study how these hidden parameters affect lexicase's ability to exploit gradients and maintain specialists using diagnostic metrics. By varying the population size with a fixed evaluation budget, we show that smaller populations tend to have greater exploitation capabilities, whereas larger populations tend to maintain more specialists. We also consider the effect redundant test cases have on specialist maintenance, and find that high redundancy may hinder the ability to optimize and maintain specialists, even for larger populations. Ultimately, we highlight that population size, evaluation budget, and test cases must be carefully considered for the characteristics of the problem being solved.
Lexidate: Model Evaluation and Selection with Lexicase
Jun 17, 2024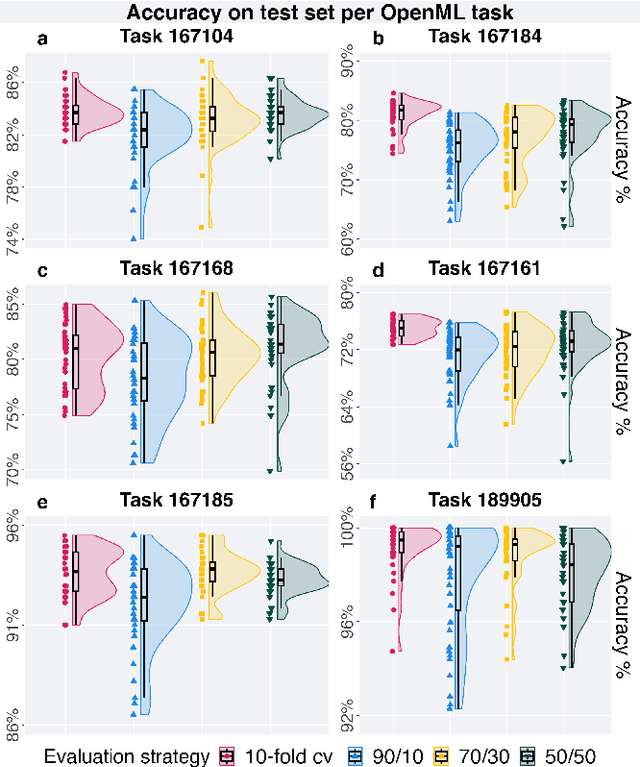
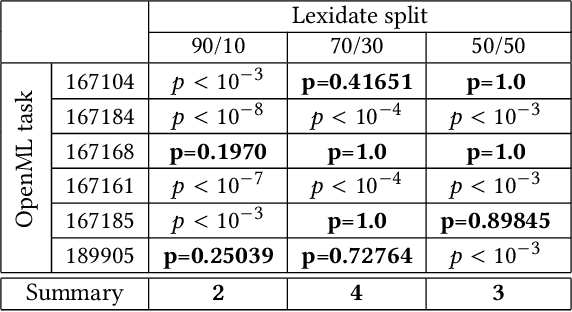
Abstract:Automated machine learning streamlines the task of finding effective machine learning pipelines by automating model training, evaluation, and selection. Traditional evaluation strategies, like cross-validation (CV), generate one value that averages the accuracy of a pipeline's predictions. This single value, however, may not fully describe the generalizability of the pipeline. Here, we present Lexicase-based Validation (lexidate), a method that uses multiple, independent prediction values for selection. Lexidate splits training data into a learning set and a selection set. Pipelines are trained on the learning set and make predictions on the selection set. The predictions are graded for correctness and used by lexicase selection to identify parent pipelines. Compared to 10-fold CV, lexicase reduces the training time. We test the effectiveness of three lexidate configurations within the Tree-based Pipeline Optimization Tool 2 (TPOT2) package on six OpenML classification tasks. In one configuration, we detected no difference in the accuracy of the final model returned from TPOT2 on most tasks compared to 10-fold CV. All configurations studied here returned similar or less complex final pipelines compared to 10-fold CV.
Phylogeny-informed fitness estimation
Jun 06, 2023



Abstract:Phylogenies (ancestry trees) depict the evolutionary history of an evolving population. In evolutionary computing, a phylogeny can reveal how an evolutionary algorithm steers a population through a search space, illuminating the step-by-step process by which any solutions evolve. Thus far, phylogenetic analyses have primarily been applied as post-hoc analyses used to deepen our understanding of existing evolutionary algorithms. Here, we investigate whether phylogenetic analyses can be used at runtime to augment parent selection procedures during an evolutionary search. Specifically, we propose phylogeny-informed fitness estimation, which exploits a population's phylogeny to estimate fitness evaluations. We evaluate phylogeny-informed fitness estimation in the context of the down-sampled lexicase and cohort lexicase selection algorithms on two diagnostic analyses and four genetic programming (GP) problems. Our results indicate that phylogeny-informed fitness estimation can mitigate the drawbacks of down-sampled lexicase, improving diversity maintenance and search space exploration. However, the extent to which phylogeny-informed fitness estimation improves problem-solving success for GP varies by problem, subsampling method, and subsampling level. This work serves as an initial step toward improving evolutionary algorithms by exploiting runtime phylogenetic analysis.
A suite of diagnostic metrics for characterizing selection schemes
Apr 29, 2022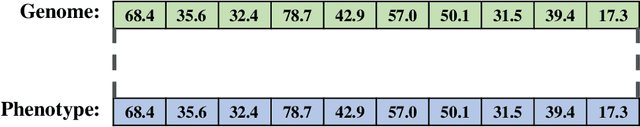
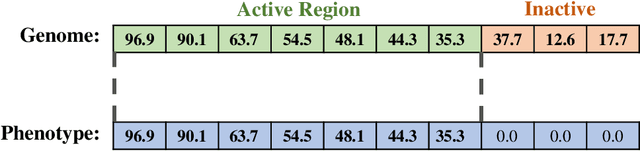
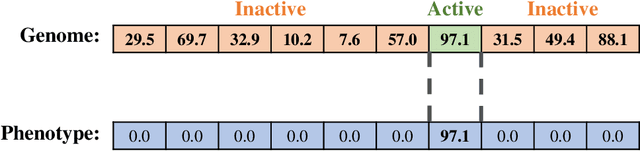
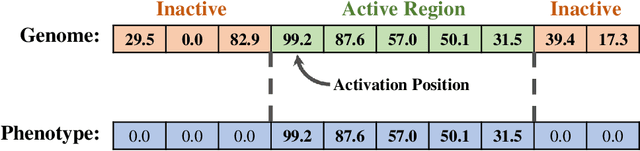
Abstract:Evolutionary algorithms are effective general-purpose techniques for solving optimization problems. Understanding how each component of an evolutionary algorithm influences its problem-solving success improves our ability to target particular problem domains. Our work focuses on evaluating selection schemes, which choose individuals to contribute genetic material to the next generation. We introduce four diagnostic search spaces for testing the strengths and weaknesses of selection schemes: the exploitation rate diagnostic, ordered exploitation rate diagnostic, contradictory objectives diagnostic, and the multi-path exploration diagnostic. Each diagnostic is handcrafted to isolate and measure the relative exploitation and exploration characteristics of selection schemes. In this study, we use our diagnostics to evaluate six population selection methods: truncation selection, tournament selection, fitness sharing, lexicase selection, nondominated sorting, and novelty search. Expectedly, tournament and truncation selection excelled in gradient exploitation but poorly explored search spaces, and novelty search excelled at exploration but failed to exploit fitness gradients. Fitness sharing performed poorly across all diagnostics, suggesting poor overall exploitation and exploration abilities. Nondominated sorting was best for maintaining populations comprised of individuals with different trade-offs of multiple objectives, but struggled to effectively exploit fitness gradients. Lexicase selection balanced search space exploration with exploitation, generally performing well across diagnostics. Our work demonstrates the value of diagnostic search spaces for building a deeper understanding of selection schemes, which can then be used to improve or develop new selection methods.
What can phylogenetic metrics tell us about useful diversity in evolutionary algorithms?
Aug 28, 2021
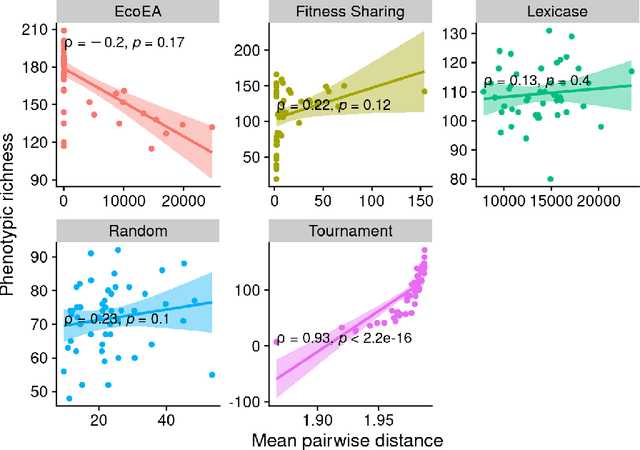
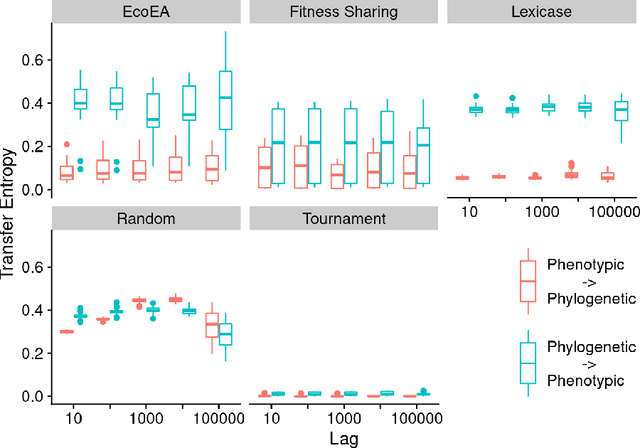
Abstract:It is generally accepted that "diversity" is associated with success in evolutionary algorithms. However, diversity is a broad concept that can be measured and defined in a multitude of ways. To date, most evolutionary computation research has measured diversity using the richness and/or evenness of a particular genotypic or phenotypic property. While these metrics are informative, we hypothesize that other diversity metrics are more strongly predictive of success. Phylogenetic diversity metrics are a class of metrics popularly used in biology, which take into account the evolutionary history of a population. Here, we investigate the extent to which 1) these metrics provide different information than those traditionally used in evolutionary computation, and 2) these metrics better predict the long-term success of a run of evolutionary computation. We find that, in most cases, phylogenetic metrics behave meaningfully differently from other diversity metrics. Moreover, our results suggest that phylogenetic diversity is indeed a better predictor of success.
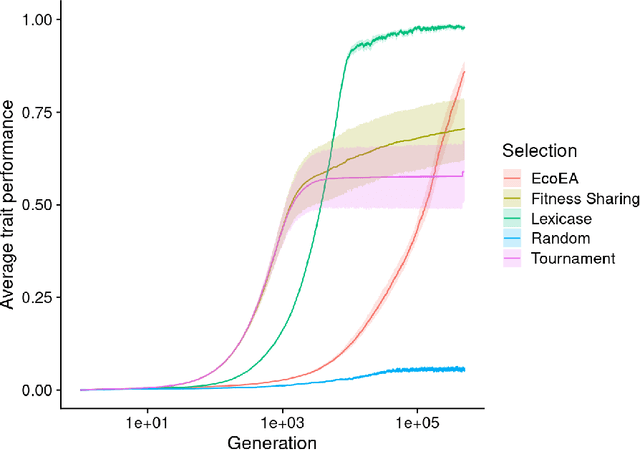
An Exploration of Exploration: Measuring the ability of lexicase selection to find obscure pathways to optimality
Jul 26, 2021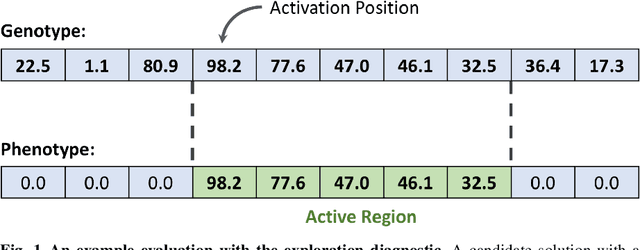
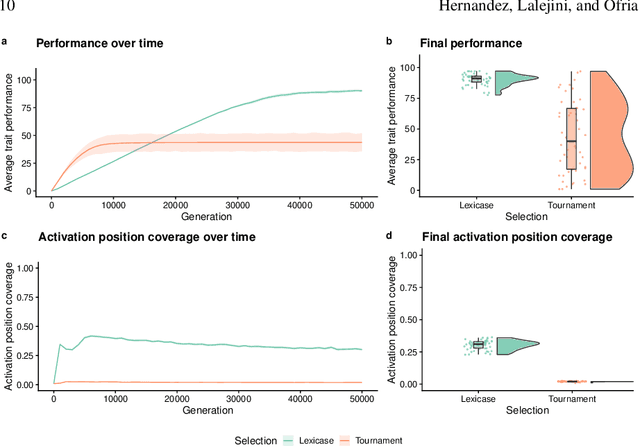
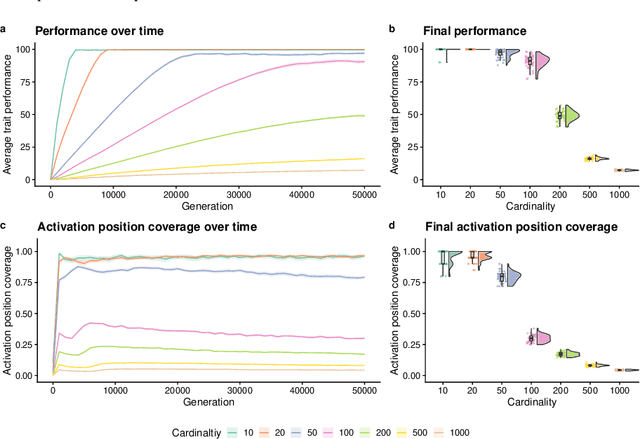
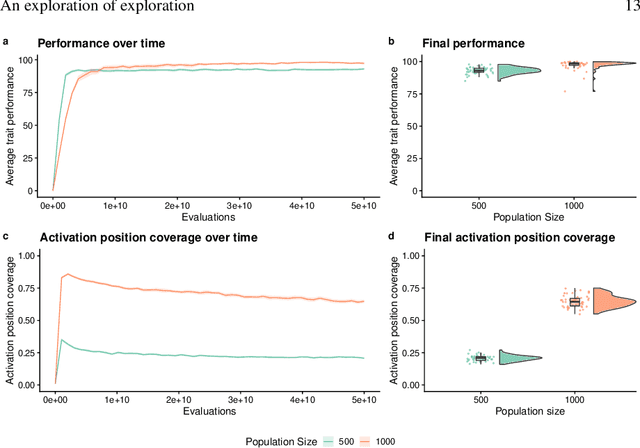
Abstract:Parent selection algorithms (selection schemes) steer populations through a problem's search space, often trading off between exploitation and exploration. Understanding how selection schemes affect exploitation and exploration within a search space is crucial to tackling increasingly challenging problems. Here, we introduce an "exploration diagnostic" that diagnoses a selection scheme's capacity for search space exploration. We use our exploration diagnostic to investigate the exploratory capacity of lexicase selection and several of its variants: epsilon lexicase, down-sampled lexicase, cohort lexicase, and novelty-lexicase. We verify that lexicase selection out-explores tournament selection, and we show that lexicase selection's exploratory capacity can be sensitive to the ratio between population size and the number of test cases used for evaluating candidate solutions. Additionally, we find that relaxing lexicase's elitism with epsilon lexicase can further improve exploration. Both down-sampling and cohort lexicase -- two techniques for applying random subsampling to test cases -- degrade lexicase's exploratory capacity; however, we find that cohort partitioning better preserves lexicase's exploratory capacity than down-sampling. Finally, we find evidence that novelty-lexicase's addition of novelty test cases can degrade lexicase's capacity for exploration. Overall, our findings provide hypotheses for further exploration and actionable insights and recommendations for using lexicase selection. Additionally, this work demonstrates the value of selection scheme diagnostics as a complement to more conventional benchmarking approaches to selection scheme analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge