Shreyas N. Samaga
Indian Institute of Technology Delhi
HalluZig: Hallucination Detection using Zigzag Persistence
Jan 04, 2026Abstract:The factual reliability of Large Language Models (LLMs) remains a critical barrier to their adoption in high-stakes domains due to their propensity to hallucinate. Current detection methods often rely on surface-level signals from the model's output, overlooking the failures that occur within the model's internal reasoning process. In this paper, we introduce a new paradigm for hallucination detection by analyzing the dynamic topology of the evolution of model's layer-wise attention. We model the sequence of attention matrices as a zigzag graph filtration and use zigzag persistence, a tool from Topological Data Analysis, to extract a topological signature. Our core hypothesis is that factual and hallucinated generations exhibit distinct topological signatures. We validate our framework, HalluZig, on multiple benchmarks, demonstrating that it outperforms strong baselines. Furthermore, our analysis reveals that these topological signatures are generalizable across different models and hallucination detection is possible only using structural signatures from partial network depth.
Quasi Zigzag Persistence: A Topological Framework for Analyzing Time-Varying Data
Feb 22, 2025Abstract:In this paper, we propose Quasi Zigzag Persistent Homology (QZPH) as a framework for analyzing time-varying data by integrating multiparameter persistence and zigzag persistence. To this end, we introduce a stable topological invariant that captures both static and dynamic features at different scales. We present an algorithm to compute this invariant efficiently. We show that it enhances the machine learning models when applied to tasks such as sleep-stage detection, demonstrating its effectiveness in capturing the evolving patterns in time-evolving datasets.
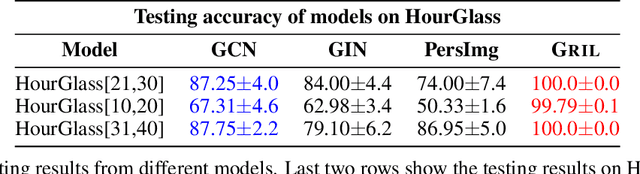
D-GRIL: End-to-End Topological Learning with 2-parameter Persistence
Jun 11, 2024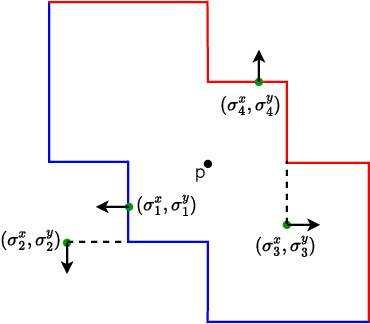
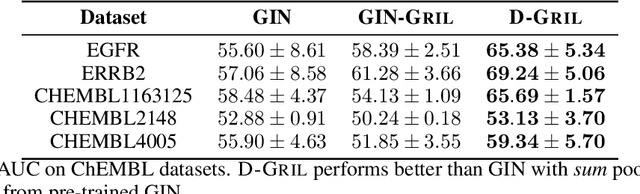
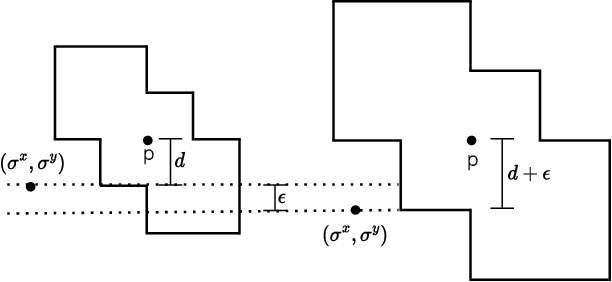
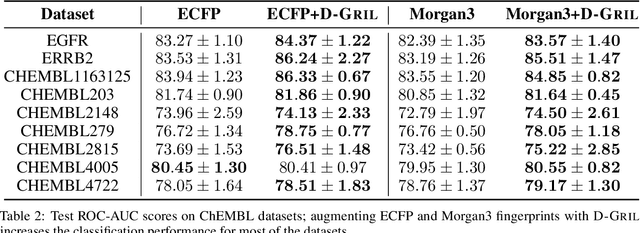
Abstract:End-to-end topological learning using 1-parameter persistence is well-known. We show that the framework can be enhanced using 2-parameter persistence by adopting a recently introduced 2-parameter persistence based vectorization technique called GRIL. We establish a theoretical foundation of differentiating GRIL producing D-GRIL. We show that D-GRIL can be used to learn a bifiltration function on standard benchmark graph datasets. Further, we exhibit that this framework can be applied in the context of bio-activity prediction in drug discovery.
TopoX: A Suite of Python Packages for Machine Learning on Topological Domains
Feb 07, 2024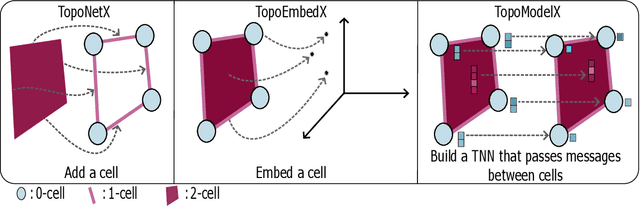
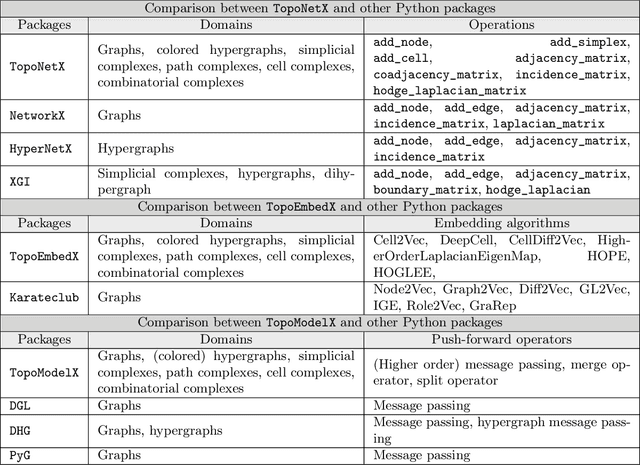
Abstract:We introduce topox, a Python software suite that provides reliable and user-friendly building blocks for computing and machine learning on topological domains that extend graphs: hypergraphs, simplicial, cellular, path and combinatorial complexes. topox consists of three packages: toponetx facilitates constructing and computing on these domains, including working with nodes, edges and higher-order cells; topoembedx provides methods to embed topological domains into vector spaces, akin to popular graph-based embedding algorithms such as node2vec; topomodelx is built on top of PyTorch and offers a comprehensive toolbox of higher-order message passing functions for neural networks on topological domains. The extensively documented and unit-tested source code of topox is available under MIT license at https://github.com/pyt-team.
ICML 2023 Topological Deep Learning Challenge : Design and Results
Oct 02, 2023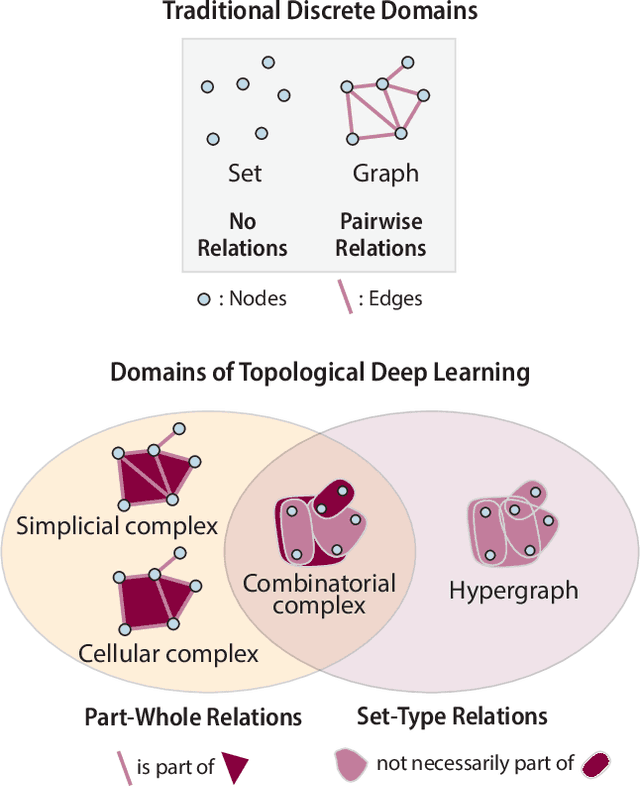
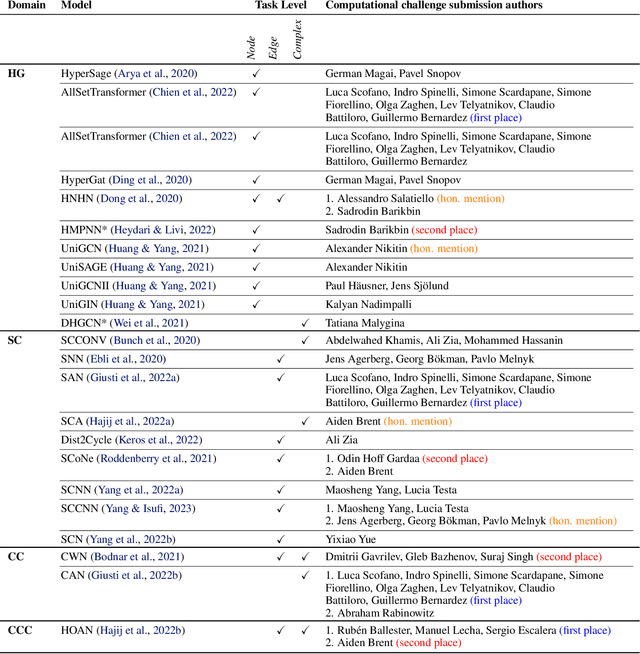
Abstract:This paper presents the computational challenge on topological deep learning that was hosted within the ICML 2023 Workshop on Topology and Geometry in Machine Learning. The competition asked participants to provide open-source implementations of topological neural networks from the literature by contributing to the python packages TopoNetX (data processing) and TopoModelX (deep learning). The challenge attracted twenty-eight qualifying submissions in its two-month duration. This paper describes the design of the challenge and summarizes its main findings.
GRIL: A $2$-parameter Persistence Based Vectorization for Machine Learning
Apr 11, 2023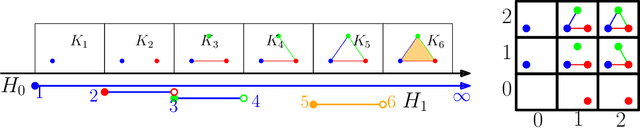
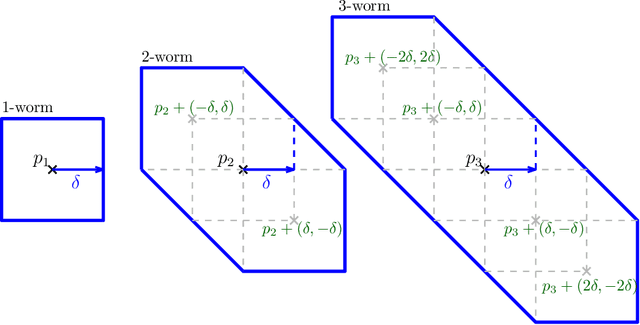
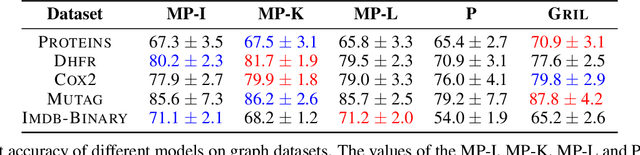
Abstract:$1$-parameter persistent homology, a cornerstone in Topological Data Analysis (TDA), studies the evolution of topological features such as connected components and cycles hidden in data. It has been applied to enhance the representation power of deep learning models, such as Graph Neural Networks (GNNs). To enrich the representations of topological features, here we propose to study $2$-parameter persistence modules induced by bi-filtration functions. In order to incorporate these representations into machine learning models, we introduce a novel vector representation called Generalized Rank Invariant Landscape \textsc{Gril} for $2$-parameter persistence modules. We show that this vector representation is $1$-Lipschitz stable and differentiable with respect to underlying filtration functions and can be easily integrated into machine learning models to augment encoding topological features. We present an algorithm to compute the vector representation efficiently. We also test our methods on synthetic and benchmark graph datasets, and compare the results with previous vector representations of $1$-parameter and $2$-parameter persistence modules.
Challenges in the application of a mortality prediction model for COVID-19 patients on an Indian cohort
Jan 15, 2021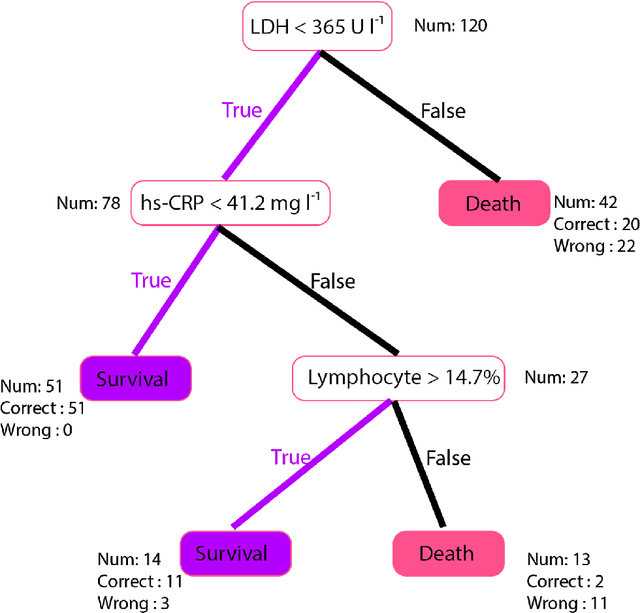
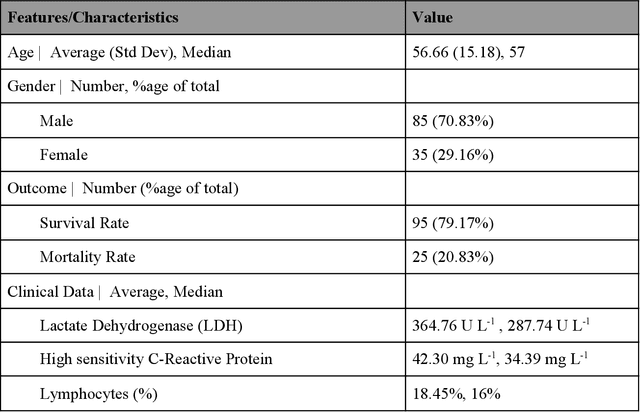
Abstract:Many countries are now experiencing the third wave of the COVID-19 pandemic straining the healthcare resources with an acute shortage of hospital beds and ventilators for the critically ill patients. This situation is especially worse in India with the second largest load of COVID-19 cases and a relatively resource-scarce medical infrastructure. Therefore, it becomes essential to triage the patients based on the severity of their disease and devote resources towards critically ill patients. Yan et al. 1 have published a very pertinent research that uses Machine learning (ML) methods to predict the outcome of COVID-19 patients based on their clinical parameters at the day of admission. They used the XGBoost algorithm, a type of ensemble model, to build the mortality prediction model. The final classifier is built through the sequential addition of multiple weak classifiers. The clinically operable decision rule was obtained from a 'single-tree XGBoost' and used lactic dehydrogenase (LDH), lymphocyte and high-sensitivity C-reactive protein (hs-CRP) values. This decision tree achieved a 100% survival prediction and 81% mortality prediction. However, these models have several technical challenges and do not provide an out of the box solution that can be deployed for other populations as has been reported in the "Matters Arising" section of Yan et al. Here, we show the limitations of this model by deploying it on one of the largest datasets of COVID-19 patients containing detailed clinical parameters collected from India.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge