Samarth Bhatia
Indian Institute of Technology, Delhi
Modularity aided consistent attributed graph clustering via coarsening
Jul 09, 2024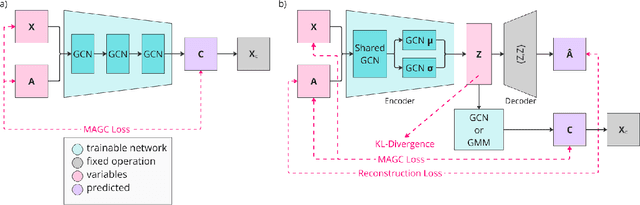
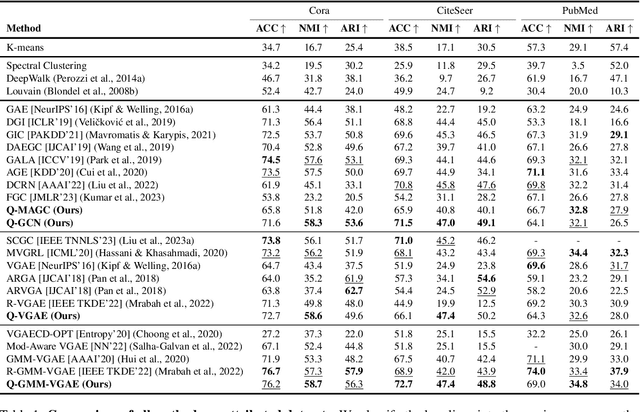
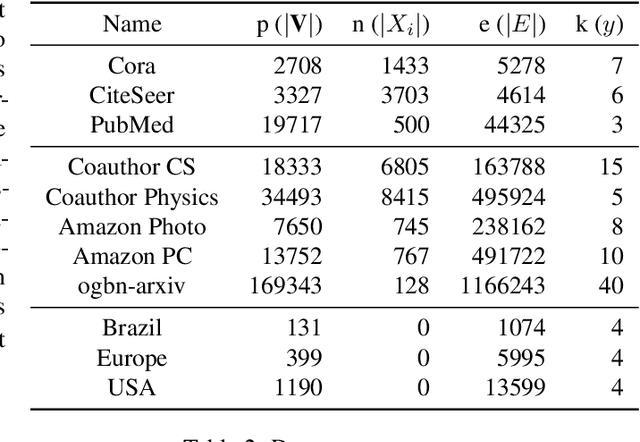
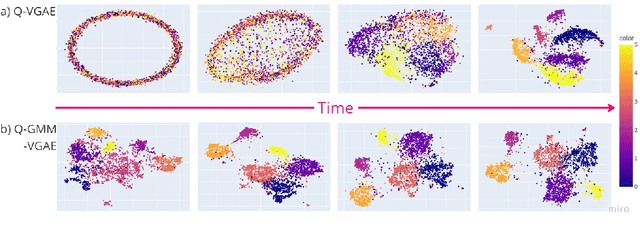
Abstract:Graph clustering is an important unsupervised learning technique for partitioning graphs with attributes and detecting communities. However, current methods struggle to accurately capture true community structures and intra-cluster relations, be computationally efficient, and identify smaller communities. We address these challenges by integrating coarsening and modularity maximization, effectively leveraging both adjacency and node features to enhance clustering accuracy. We propose a loss function incorporating log-determinant, smoothness, and modularity components using a block majorization-minimization technique, resulting in superior clustering outcomes. The method is theoretically consistent under the Degree-Corrected Stochastic Block Model (DC-SBM), ensuring asymptotic error-free performance and complete label recovery. Our provably convergent and time-efficient algorithm seamlessly integrates with graph neural networks (GNNs) and variational graph autoencoders (VGAEs) to learn enhanced node features and deliver exceptional clustering performance. Extensive experiments on benchmark datasets demonstrate its superiority over existing state-of-the-art methods for both attributed and non-attributed graphs.
Lowering PyTorch's Memory Consumption for Selective Differentiation
Apr 15, 2024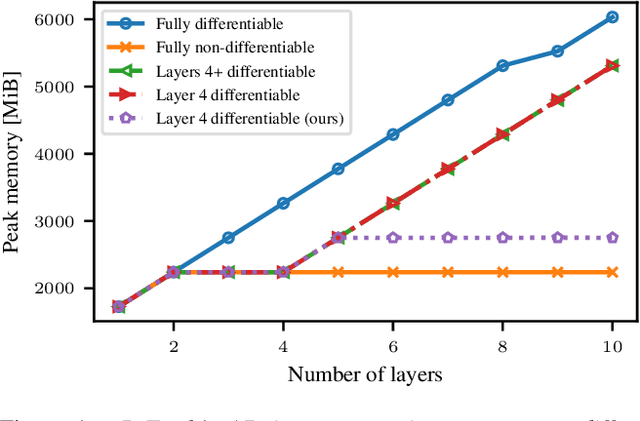
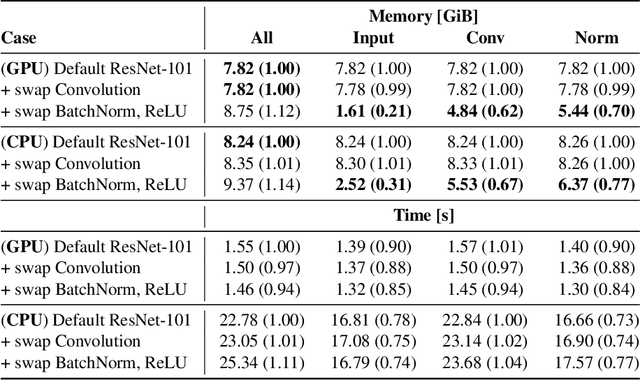
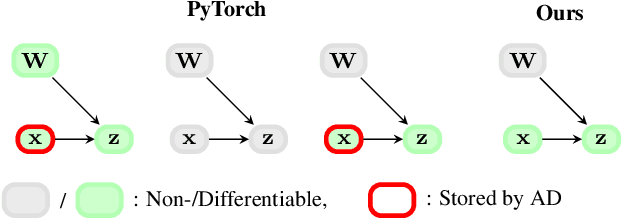

Abstract:Memory is a limiting resource for many deep learning tasks. Beside the neural network weights, one main memory consumer is the computation graph built up by automatic differentiation (AD) for backpropagation. We observe that PyTorch's current AD implementation neglects information about parameter differentiability when storing the computation graph. This information is useful though to reduce memory whenever gradients are requested for a parameter subset, as is the case in many modern fine-tuning tasks. Specifically, inputs to layers that act linearly in their parameters (dense, convolution, or normalization layers) can be discarded whenever the parameters are marked as non-differentiable. We provide a drop-in, differentiability-agnostic implementation of such layers and demonstrate its ability to reduce memory without affecting run time.
Severity and Mortality Prediction Models to Triage Indian COVID-19 Patients
Sep 02, 2021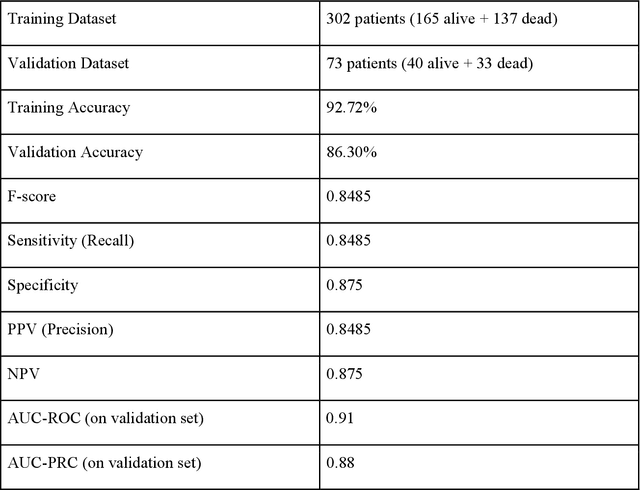
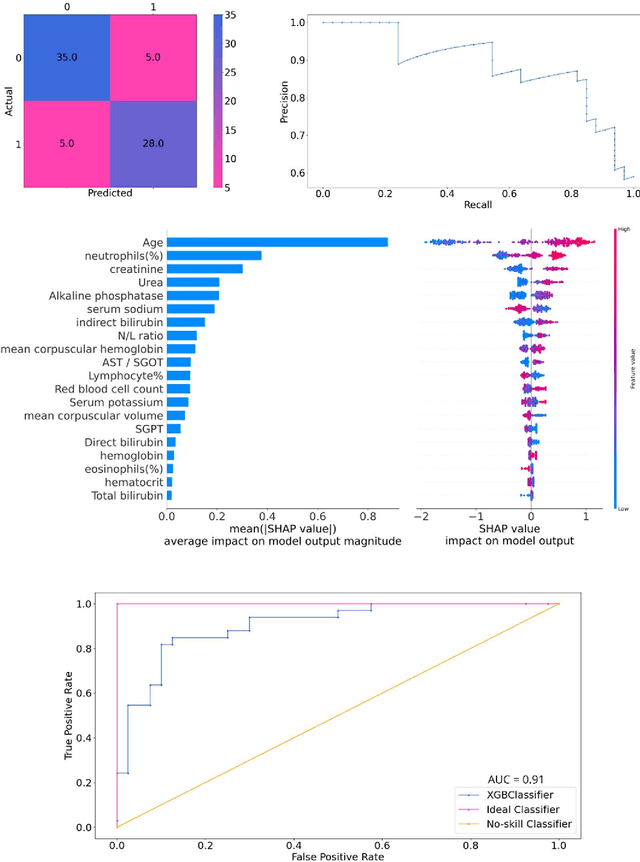
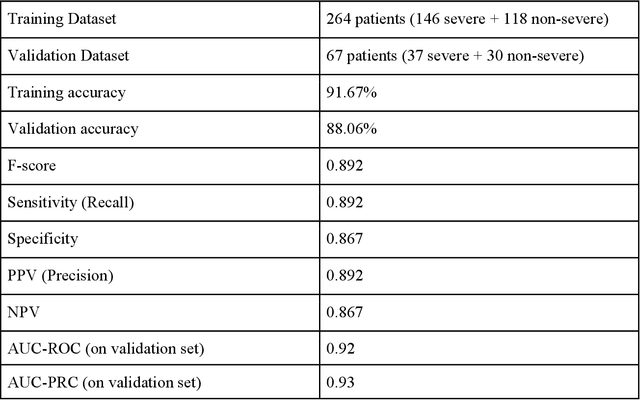
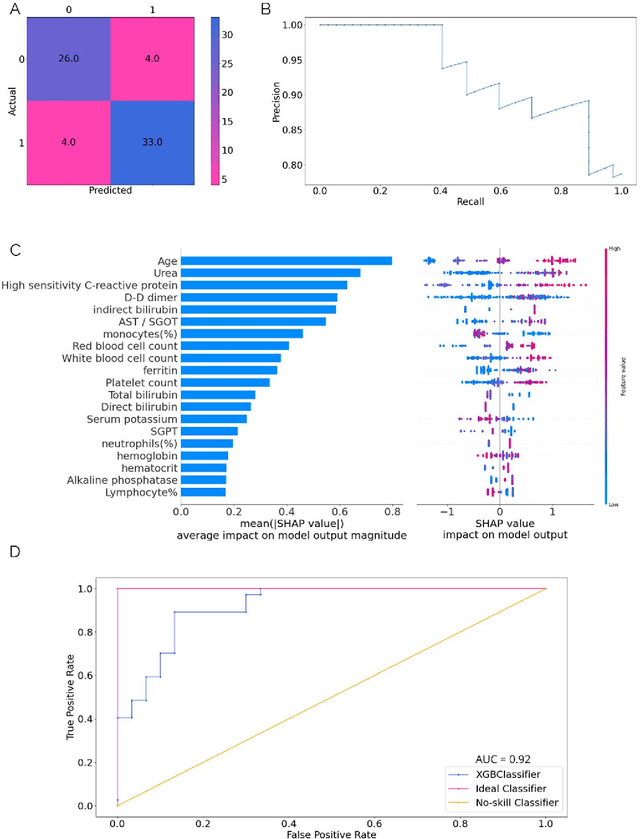
Abstract:As the second wave in India mitigates, COVID-19 has now infected about 29 million patients countrywide, leading to more than 350 thousand people dead. As the infections surged, the strain on the medical infrastructure in the country became apparent. While the country vaccinates its population, opening up the economy may lead to an increase in infection rates. In this scenario, it is essential to effectively utilize the limited hospital resources by an informed patient triaging system based on clinical parameters. Here, we present two interpretable machine learning models predicting the clinical outcomes, severity, and mortality, of the patients based on routine non-invasive surveillance of blood parameters from one of the largest cohorts of Indian patients at the day of admission. Patient severity and mortality prediction models achieved 86.3% and 88.06% accuracy, respectively, with an AUC-ROC of 0.91 and 0.92. We have integrated both the models in a user-friendly web app calculator, https://triage-COVID-19.herokuapp.com/, to showcase the potential deployment of such efforts at scale.
Challenges in the application of a mortality prediction model for COVID-19 patients on an Indian cohort
Jan 15, 2021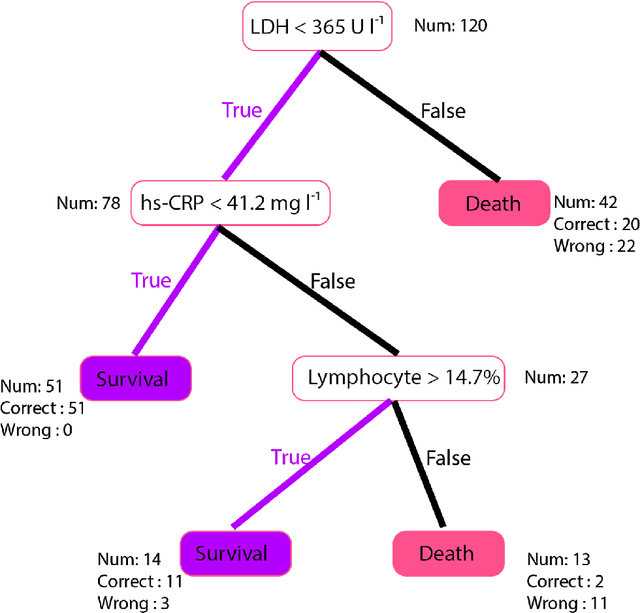
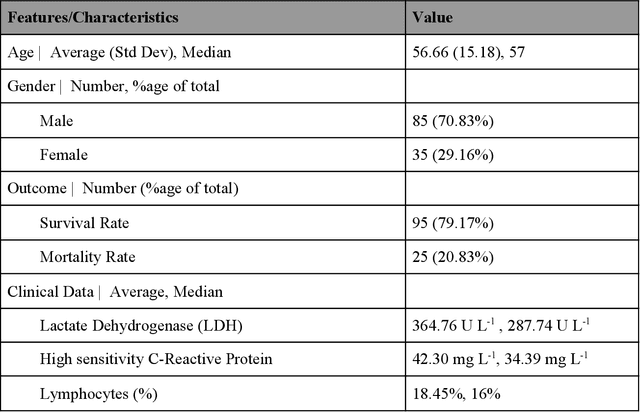
Abstract:Many countries are now experiencing the third wave of the COVID-19 pandemic straining the healthcare resources with an acute shortage of hospital beds and ventilators for the critically ill patients. This situation is especially worse in India with the second largest load of COVID-19 cases and a relatively resource-scarce medical infrastructure. Therefore, it becomes essential to triage the patients based on the severity of their disease and devote resources towards critically ill patients. Yan et al. 1 have published a very pertinent research that uses Machine learning (ML) methods to predict the outcome of COVID-19 patients based on their clinical parameters at the day of admission. They used the XGBoost algorithm, a type of ensemble model, to build the mortality prediction model. The final classifier is built through the sequential addition of multiple weak classifiers. The clinically operable decision rule was obtained from a 'single-tree XGBoost' and used lactic dehydrogenase (LDH), lymphocyte and high-sensitivity C-reactive protein (hs-CRP) values. This decision tree achieved a 100% survival prediction and 81% mortality prediction. However, these models have several technical challenges and do not provide an out of the box solution that can be deployed for other populations as has been reported in the "Matters Arising" section of Yan et al. Here, we show the limitations of this model by deploying it on one of the largest datasets of COVID-19 patients containing detailed clinical parameters collected from India.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge