Seonwoo Min
Transformers meet Stochastic Block Models: Attention with Data-Adaptive Sparsity and Cost
Oct 27, 2022



Abstract:To overcome the quadratic cost of self-attention, recent works have proposed various sparse attention modules, most of which fall under one of two groups: 1) sparse attention under a hand-crafted patterns and 2) full attention followed by a sparse variant of softmax such as $\alpha$-entmax. Unfortunately, the first group lacks adaptability to data while the second still requires quadratic cost in training. In this work, we propose SBM-Transformer, a model that resolves both problems by endowing each attention head with a mixed-membership Stochastic Block Model (SBM). Then, each attention head data-adaptively samples a bipartite graph, the adjacency of which is used as an attention mask for each input. During backpropagation, a straight-through estimator is used to flow gradients beyond the discrete sampling step and adjust the probabilities of sampled edges based on the predictive loss. The forward and backward cost are thus linear to the number of edges, which each attention head can also choose flexibly based on the input. By assessing the distribution of graphs, we theoretically show that SBM-Transformer is a universal approximator for arbitrary sequence-to-sequence functions in expectation. Empirical evaluations under the LRA and GLUE benchmarks demonstrate that our model outperforms previous efficient variants as well as the original Transformer with full attention. Our implementation can be found in https://github.com/sc782/SBM-Transformer .
Grounding Visual Representations with Texts for Domain Generalization
Jul 21, 2022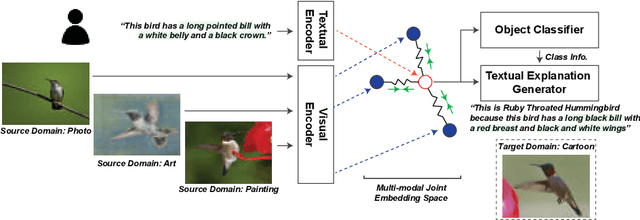
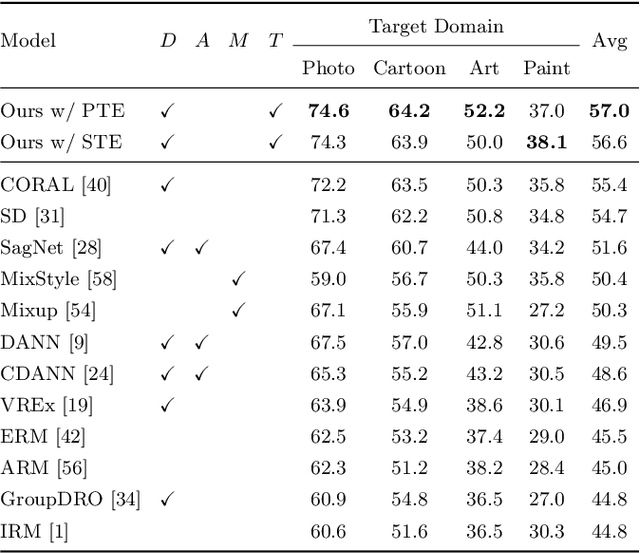
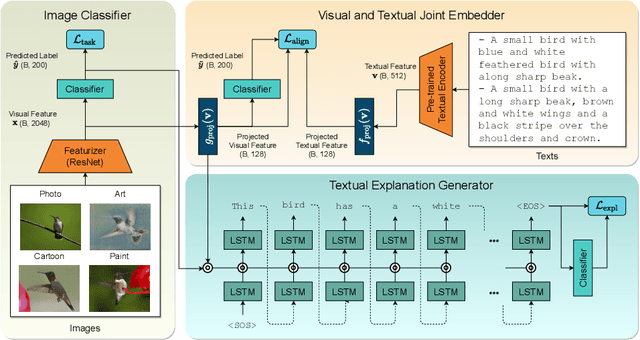
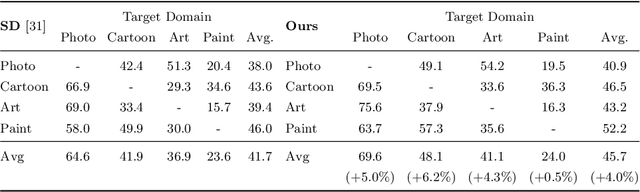
Abstract:Reducing the representational discrepancy between source and target domains is a key component to maximize the model generalization. In this work, we advocate for leveraging natural language supervision for the domain generalization task. We introduce two modules to ground visual representations with texts containing typical reasoning of humans: (1) Visual and Textual Joint Embedder and (2) Textual Explanation Generator. The former learns the image-text joint embedding space where we can ground high-level class-discriminative information into the model. The latter leverages an explainable model and generates explanations justifying the rationale behind its decision. To the best of our knowledge, this is the first work to leverage the vision-and-language cross-modality approach for the domain generalization task. Our experiments with a newly created CUB-DG benchmark dataset demonstrate that cross-modality supervision can be successfully used to ground domain-invariant visual representations and improve the model generalization. Furthermore, in the large-scale DomainBed benchmark, our proposed method achieves state-of-the-art results and ranks 1st in average performance for five multi-domain datasets. The dataset and codes are available at https://github.com/mswzeus/GVRT.
Pure Transformers are Powerful Graph Learners
Jul 06, 2022



Abstract:We show that standard Transformers without graph-specific modifications can lead to promising results in graph learning both in theory and practice. Given a graph, we simply treat all nodes and edges as independent tokens, augment them with token embeddings, and feed them to a Transformer. With an appropriate choice of token embeddings, we prove that this approach is theoretically at least as expressive as an invariant graph network (2-IGN) composed of equivariant linear layers, which is already more expressive than all message-passing Graph Neural Networks (GNN). When trained on a large-scale graph dataset (PCQM4Mv2), our method coined Tokenized Graph Transformer (TokenGT) achieves significantly better results compared to GNN baselines and competitive results compared to Transformer variants with sophisticated graph-specific inductive bias. Our implementation is available at https://github.com/jw9730/tokengt.
Supervised Neural Discrete Universal Denoiser for Adaptive Denoising
Nov 24, 2021



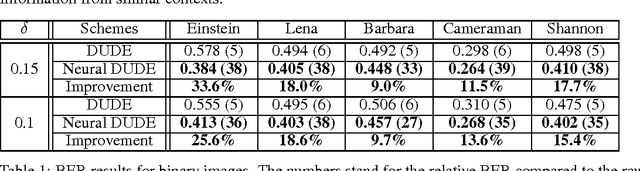
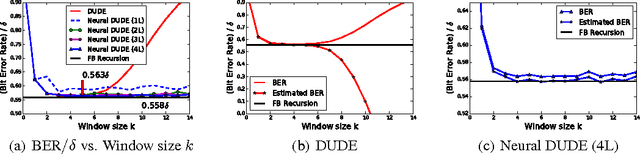
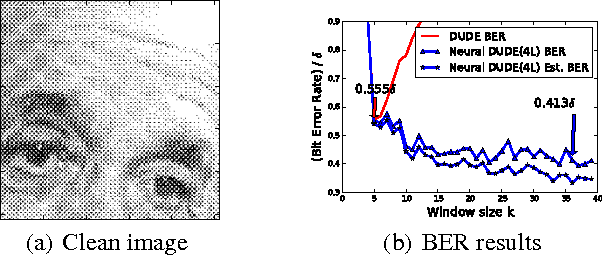
Abstract:We improve the recently developed Neural DUDE, a neural network-based adaptive discrete denoiser, by combining it with the supervised learning framework. Namely, we make the supervised pre-training of Neural DUDE compatible with the adaptive fine-tuning of the parameters based on the given noisy data subject to denoising. As a result, we achieve a significant denoising performance boost compared to the vanilla Neural DUDE, which only carries out the adaptive fine-tuning step with randomly initialized parameters. Moreover, we show the adaptive fine-tuning makes the algorithm robust such that a noise-mismatched or blindly trained supervised model can still achieve the performance of that of the matched model. Furthermore, we make a few algorithmic advancements to make Neural DUDE more scalable and deal with multi-dimensional data or data with larger alphabet size. We systematically show our improvements on two very diverse datasets, binary images and DNA sequences.
TargetNet: Functional microRNA Target Prediction with Deep Neural Networks
Jul 23, 2021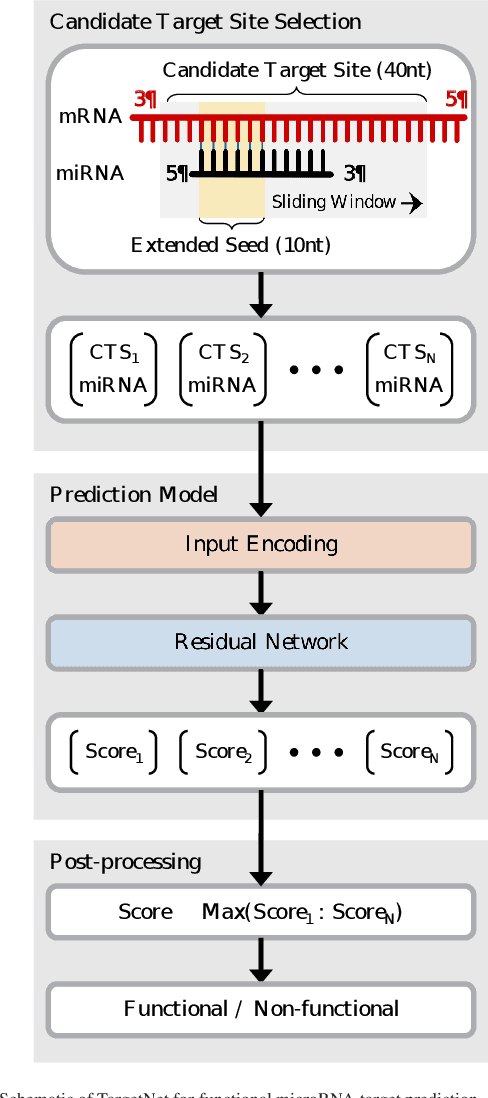
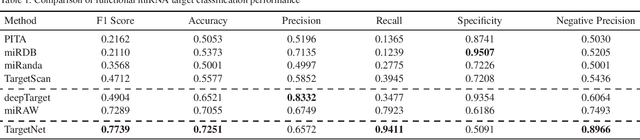
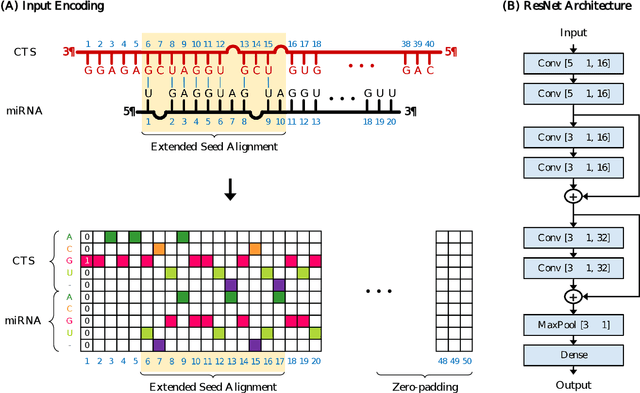
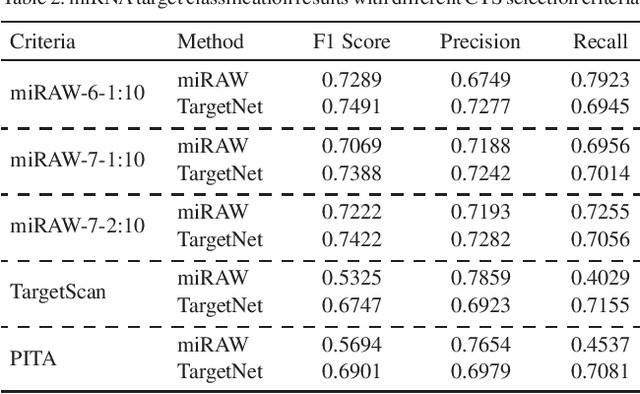
Abstract:MicroRNAs (miRNAs) play pivotal roles in gene expression regulation by binding to target sites of messenger RNAs (mRNAs). While identifying functional targets of miRNAs is of utmost importance, their prediction remains a great challenge. Previous computational algorithms have major limitations. They use conservative candidate target site (CTS) selection criteria mainly focusing on canonical site types, rely on laborious and time-consuming manual feature extraction, and do not fully capitalize on the information underlying miRNA-CTS interactions. In this paper, we introduce TargetNet, a novel deep learning-based algorithm for functional miRNA target prediction. To address the limitations of previous approaches, TargetNet has three key components: (1) relaxed CTS selection criteria accommodating irregularities in the seed region, (2) a novel miRNA-CTS sequence encoding scheme incorporating extended seed region alignments, and (3) a deep residual network-based prediction model. The proposed model was trained with miRNA-CTS pair datasets and evaluated with miRNA-mRNA pair datasets. TargetNet advances the previous state-of-the-art algorithms used in functional miRNA target classification. Furthermore, it demonstrates great potential for distinguishing high-functional miRNA targets.
Pre-Training of Deep Bidirectional Protein Sequence Representations with Structural Information
Nov 25, 2019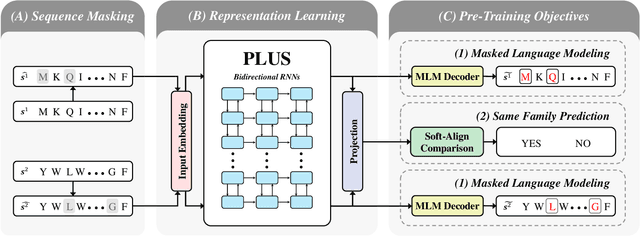
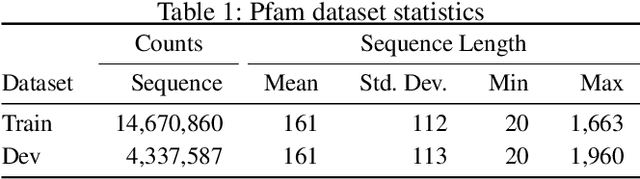
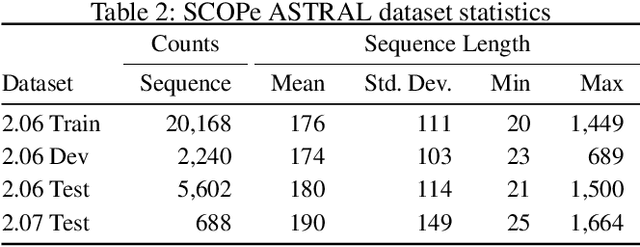
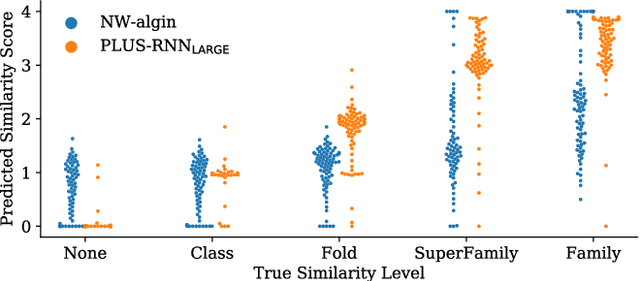
Abstract:A structure of a protein has a direct impact on its properties and functions. However, identification of structural similarity directly from amino acid sequences remains as a challenging problem in computational biology. In this paper, we introduce a novel BERT-wise pre-training scheme for a protein sequence representation model called PLUS, which stands for Protein sequence representations Learned Using Structural information. As natural language representation models capture syntactic and semantic information of words from a large unlabeled text corpus, PLUS captures structural information of amino acids from a large weakly labeled protein database. Since the Transformer encoder, BERT's original model architecture, has a severe computational requirement to handle long sequences, we first propose to combine a bidirectional recurrent neural network with the BERT-wise pre-training scheme. PLUS is designed to learn protein representations with two pre-training objectives, i.e., masked language modeling and same family prediction. Then, the pre-trained model can be fine-tuned for a wide range of tasks without training randomly initialized task-specific models from scratch. It obtains new state-of-the-art results on both (1) protein-level and (2) amino-acid-level tasks, outperforming many task-specific algorithms.
Neural Universal Discrete Denoiser
Aug 24, 2016
Abstract:We present a new framework of applying deep neural networks (DNN) to devise a universal discrete denoiser. Unlike other approaches that utilize supervised learning for denoising, we do not require any additional training data. In such setting, while the ground-truth label, i.e., the clean data, is not available, we devise "pseudo-labels" and a novel objective function such that DNN can be trained in a same way as supervised learning to become a discrete denoiser. We experimentally show that our resulting algorithm, dubbed as Neural DUDE, significantly outperforms the previous state-of-the-art in several applications with a systematic rule of choosing the hyperparameter, which is an attractive feature in practice.
Deep Learning in Bioinformatics
Jun 19, 2016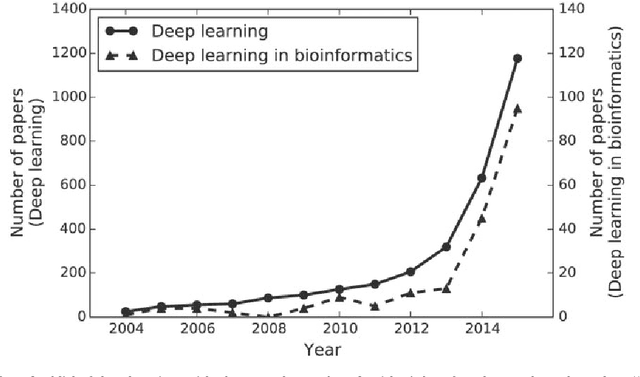
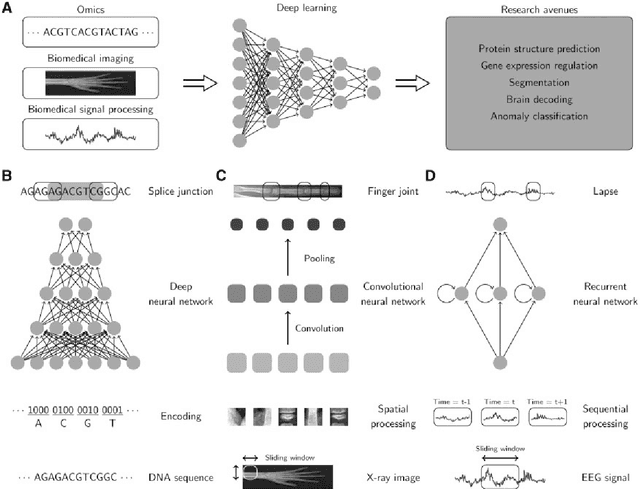
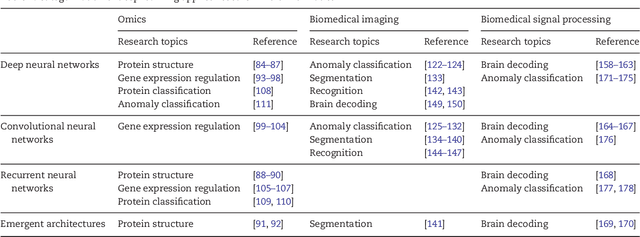
Abstract:In the era of big data, transformation of biomedical big data into valuable knowledge has been one of the most important challenges in bioinformatics. Deep learning has advanced rapidly since the early 2000s and now demonstrates state-of-the-art performance in various fields. Accordingly, application of deep learning in bioinformatics to gain insight from data has been emphasized in both academia and industry. Here, we review deep learning in bioinformatics, presenting examples of current research. To provide a useful and comprehensive perspective, we categorize research both by the bioinformatics domain (i.e., omics, biomedical imaging, biomedical signal processing) and deep learning architecture (i.e., deep neural networks, convolutional neural networks, recurrent neural networks, emergent architectures) and present brief descriptions of each study. Additionally, we discuss theoretical and practical issues of deep learning in bioinformatics and suggest future research directions. We believe that this review will provide valuable insights and serve as a starting point for researchers to apply deep learning approaches in their bioinformatics studies.
deepMiRGene: Deep Neural Network based Precursor microRNA Prediction
Apr 29, 2016



Abstract:Since microRNAs (miRNAs) play a crucial role in post-transcriptional gene regulation, miRNA identification is one of the most essential problems in computational biology. miRNAs are usually short in length ranging between 20 and 23 base pairs. It is thus often difficult to distinguish miRNA-encoding sequences from other non-coding RNAs and pseudo miRNAs that have a similar length, and most previous studies have recommended using precursor miRNAs instead of mature miRNAs for robust detection. A great number of conventional machine-learning-based classification methods have been proposed, but they often have the serious disadvantage of requiring manual feature engineering, and their performance is limited as well. In this paper, we propose a novel miRNA precursor prediction algorithm, deepMiRGene, based on recurrent neural networks, specifically long short-term memory networks. deepMiRGene automatically learns suitable features from the data themselves without manual feature engineering and constructs a model that can successfully reflect structural characteristics of precursor miRNAs. For the performance evaluation of our approach, we have employed several widely used evaluation metrics on three recent benchmark datasets and verified that deepMiRGene delivered comparable performance among the current state-of-the-art tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge